
Trypanosoma vivax (strain Y486)
Taxonomy: cellular organisms; Eukaryota; Discoba; Euglenozoa; Kinetoplastea; Metakinetoplastina; Trypanosomatida; Trypanosomatidae; Trypanosoma; Duttonella; Trypanosoma vivax
Average proteome isoelectric point is 7.44
Get precalculated fractions of proteins
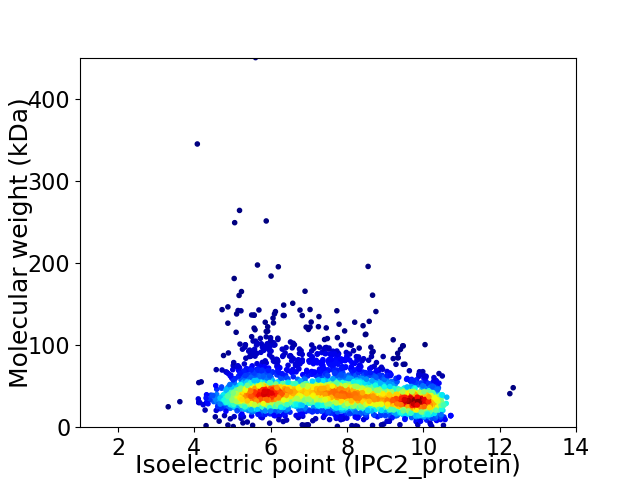
Virtual 2D-PAGE plot for 3778 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F9WTW1|F9WTW1_TRYVY Uncharacterized protein OS=Trypanosoma vivax (strain Y486) OX=1055687 GN=TvY486_0039010 PE=4 SV=1
MM1 pKa = 7.92LDD3 pKa = 3.98LSHH6 pKa = 6.77CTGITDD12 pKa = 3.97VSPLTTLIEE21 pKa = 4.43LKK23 pKa = 10.76EE24 pKa = 4.19LDD26 pKa = 5.02LNDD29 pKa = 3.59CTGITDD35 pKa = 3.8VSPLSTLIRR44 pKa = 11.84LEE46 pKa = 3.92KK47 pKa = 10.87LNLSDD52 pKa = 3.8CTGITDD58 pKa = 3.92VSPLSTLIRR67 pKa = 11.84LEE69 pKa = 3.79KK70 pKa = 10.79LNLSGCTGITDD81 pKa = 3.76VSPLTTLIEE90 pKa = 4.43LKK92 pKa = 10.76EE93 pKa = 4.19LDD95 pKa = 4.98LNDD98 pKa = 3.6CTRR101 pKa = 11.84ITDD104 pKa = 3.65VSPLSTLIRR113 pKa = 11.84LEE115 pKa = 3.98KK116 pKa = 10.62LCLSGCTGITDD127 pKa = 3.76VSPLTTLIEE136 pKa = 4.21LKK138 pKa = 10.07EE139 pKa = 4.15LCLSGCTGITDD150 pKa = 3.76VSPLTTLIEE159 pKa = 4.21LKK161 pKa = 10.7EE162 pKa = 4.26LGLSGCTGITDD173 pKa = 3.76VSPLTTLIEE182 pKa = 4.21LKK184 pKa = 10.7EE185 pKa = 4.26LGLSGCTGITDD196 pKa = 3.76VSPLTTLIRR205 pKa = 11.84LKK207 pKa = 10.53VLYY210 pKa = 10.59LIGCTGITDD219 pKa = 3.8VSPLTTLIEE228 pKa = 4.24LKK230 pKa = 10.72EE231 pKa = 4.2LDD233 pKa = 3.97LHH235 pKa = 6.69DD236 pKa = 4.19CTGITDD242 pKa = 3.87VSPLTTLIEE251 pKa = 4.21LKK253 pKa = 10.7EE254 pKa = 4.26LGLSGCTGITDD265 pKa = 3.76VSPLTTLIRR274 pKa = 11.84LEE276 pKa = 3.81VLYY279 pKa = 10.91LIGCTGITDD288 pKa = 3.8VSPLTTLIEE297 pKa = 4.24LKK299 pKa = 10.72EE300 pKa = 4.2LDD302 pKa = 3.97LHH304 pKa = 6.69DD305 pKa = 4.19CTGITDD311 pKa = 3.87VSPLTTLIEE320 pKa = 4.29LKK322 pKa = 10.57EE323 pKa = 3.85LALYY327 pKa = 10.4GCTRR331 pKa = 11.84ITDD334 pKa = 3.77VSPLSALIRR343 pKa = 11.84LEE345 pKa = 3.9KK346 pKa = 10.7LCLSGCTGITDD357 pKa = 3.76VSPLTTLIRR366 pKa = 11.84LEE368 pKa = 3.81VLYY371 pKa = 10.91LIGCTGITDD380 pKa = 3.8VSPLTTLIEE389 pKa = 4.21LKK391 pKa = 10.7EE392 pKa = 4.26LGLSGCTGITDD403 pKa = 3.76VSPLTTLIEE412 pKa = 4.21LKK414 pKa = 10.7EE415 pKa = 4.26LGLSGCTGITDD426 pKa = 3.76VSPLTTLIRR435 pKa = 11.84LEE437 pKa = 3.81VLYY440 pKa = 10.91LIGCTGITDD449 pKa = 3.8VSPLTTLIEE458 pKa = 4.24LKK460 pKa = 10.72EE461 pKa = 4.2LDD463 pKa = 3.97LHH465 pKa = 6.66DD466 pKa = 4.18CTGISDD472 pKa = 3.94VSPLPMMIRR481 pKa = 11.84PEE483 pKa = 4.06VLYY486 pKa = 10.03MIGCTGISDD495 pKa = 3.94VSPLTTMIEE504 pKa = 4.33FKK506 pKa = 10.55GAEE509 pKa = 4.12YY510 pKa = 10.32EE511 pKa = 4.1
MM1 pKa = 7.92LDD3 pKa = 3.98LSHH6 pKa = 6.77CTGITDD12 pKa = 3.97VSPLTTLIEE21 pKa = 4.43LKK23 pKa = 10.76EE24 pKa = 4.19LDD26 pKa = 5.02LNDD29 pKa = 3.59CTGITDD35 pKa = 3.8VSPLSTLIRR44 pKa = 11.84LEE46 pKa = 3.92KK47 pKa = 10.87LNLSDD52 pKa = 3.8CTGITDD58 pKa = 3.92VSPLSTLIRR67 pKa = 11.84LEE69 pKa = 3.79KK70 pKa = 10.79LNLSGCTGITDD81 pKa = 3.76VSPLTTLIEE90 pKa = 4.43LKK92 pKa = 10.76EE93 pKa = 4.19LDD95 pKa = 4.98LNDD98 pKa = 3.6CTRR101 pKa = 11.84ITDD104 pKa = 3.65VSPLSTLIRR113 pKa = 11.84LEE115 pKa = 3.98KK116 pKa = 10.62LCLSGCTGITDD127 pKa = 3.76VSPLTTLIEE136 pKa = 4.21LKK138 pKa = 10.07EE139 pKa = 4.15LCLSGCTGITDD150 pKa = 3.76VSPLTTLIEE159 pKa = 4.21LKK161 pKa = 10.7EE162 pKa = 4.26LGLSGCTGITDD173 pKa = 3.76VSPLTTLIEE182 pKa = 4.21LKK184 pKa = 10.7EE185 pKa = 4.26LGLSGCTGITDD196 pKa = 3.76VSPLTTLIRR205 pKa = 11.84LKK207 pKa = 10.53VLYY210 pKa = 10.59LIGCTGITDD219 pKa = 3.8VSPLTTLIEE228 pKa = 4.24LKK230 pKa = 10.72EE231 pKa = 4.2LDD233 pKa = 3.97LHH235 pKa = 6.69DD236 pKa = 4.19CTGITDD242 pKa = 3.87VSPLTTLIEE251 pKa = 4.21LKK253 pKa = 10.7EE254 pKa = 4.26LGLSGCTGITDD265 pKa = 3.76VSPLTTLIRR274 pKa = 11.84LEE276 pKa = 3.81VLYY279 pKa = 10.91LIGCTGITDD288 pKa = 3.8VSPLTTLIEE297 pKa = 4.24LKK299 pKa = 10.72EE300 pKa = 4.2LDD302 pKa = 3.97LHH304 pKa = 6.69DD305 pKa = 4.19CTGITDD311 pKa = 3.87VSPLTTLIEE320 pKa = 4.29LKK322 pKa = 10.57EE323 pKa = 3.85LALYY327 pKa = 10.4GCTRR331 pKa = 11.84ITDD334 pKa = 3.77VSPLSALIRR343 pKa = 11.84LEE345 pKa = 3.9KK346 pKa = 10.7LCLSGCTGITDD357 pKa = 3.76VSPLTTLIRR366 pKa = 11.84LEE368 pKa = 3.81VLYY371 pKa = 10.91LIGCTGITDD380 pKa = 3.8VSPLTTLIEE389 pKa = 4.21LKK391 pKa = 10.7EE392 pKa = 4.26LGLSGCTGITDD403 pKa = 3.76VSPLTTLIEE412 pKa = 4.21LKK414 pKa = 10.7EE415 pKa = 4.26LGLSGCTGITDD426 pKa = 3.76VSPLTTLIRR435 pKa = 11.84LEE437 pKa = 3.81VLYY440 pKa = 10.91LIGCTGITDD449 pKa = 3.8VSPLTTLIEE458 pKa = 4.24LKK460 pKa = 10.72EE461 pKa = 4.2LDD463 pKa = 3.97LHH465 pKa = 6.66DD466 pKa = 4.18CTGISDD472 pKa = 3.94VSPLPMMIRR481 pKa = 11.84PEE483 pKa = 4.06VLYY486 pKa = 10.03MIGCTGISDD495 pKa = 3.94VSPLTTMIEE504 pKa = 4.33FKK506 pKa = 10.55GAEE509 pKa = 4.12YY510 pKa = 10.32EE511 pKa = 4.1
Molecular weight: 54.37 kDa
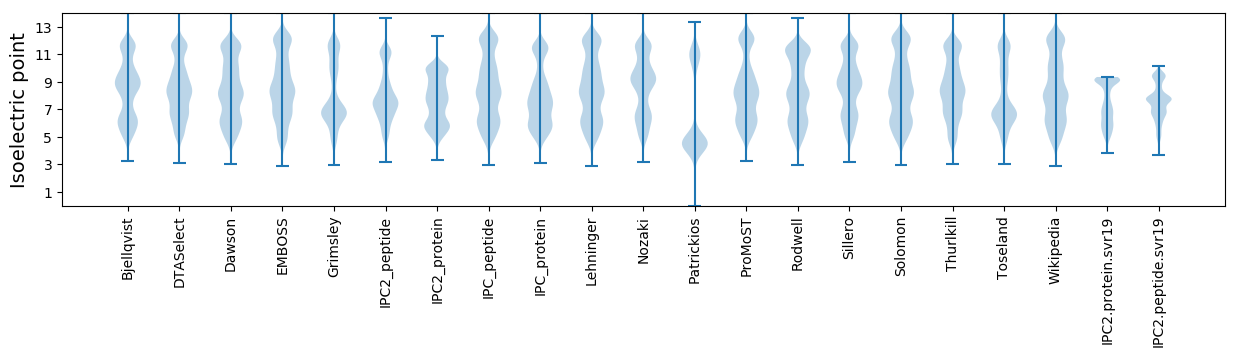
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F9WKS7|F9WKS7_TRYVY Uncharacterized protein OS=Trypanosoma vivax (strain Y486) OX=1055687 GN=TvY486_0007440 PE=4 SV=1
MM1 pKa = 7.69RR2 pKa = 11.84GKK4 pKa = 9.72HH5 pKa = 4.17VATRR9 pKa = 11.84SHH11 pKa = 5.44CHH13 pKa = 5.85NYY15 pKa = 9.83RR16 pKa = 11.84SAGQTQDD23 pKa = 3.12AQHH26 pKa = 6.63PLQRR30 pKa = 11.84RR31 pKa = 11.84AACCVSEE38 pKa = 4.56HH39 pKa = 6.15AWTVHH44 pKa = 5.06GASTRR49 pKa = 11.84TAASSACPGKK59 pKa = 8.38TLPIVHH65 pKa = 7.17AEE67 pKa = 3.79PTRR70 pKa = 11.84APRR73 pKa = 11.84CCHH76 pKa = 7.22LLCCPDD82 pKa = 3.34ASGMRR87 pKa = 11.84NADD90 pKa = 3.53TQRR93 pKa = 11.84NDD95 pKa = 3.25HH96 pKa = 6.74AVDD99 pKa = 3.48GCIFQKK105 pKa = 7.89EE106 pKa = 4.26TRR108 pKa = 11.84QPSNASAPSLAHH120 pKa = 5.75IVMVTLHH127 pKa = 6.89RR128 pKa = 11.84SCATQQTCRR137 pKa = 11.84SMRR140 pKa = 11.84APLPGHH146 pKa = 6.8LPSSGAHH153 pKa = 4.07SVQRR157 pKa = 11.84RR158 pKa = 11.84EE159 pKa = 3.78TRR161 pKa = 11.84AVAMHH166 pKa = 5.65STISTQHH173 pKa = 6.27KK174 pKa = 10.38GEE176 pKa = 4.26NNTKK180 pKa = 9.58KK181 pKa = 10.62ARR183 pKa = 11.84VRR185 pKa = 11.84HH186 pKa = 5.76LPASQSTVGQPTHH199 pKa = 5.87TRR201 pKa = 11.84TITQMCPAVTVGHH214 pKa = 6.92HH215 pKa = 6.79APPAPPPSTYY225 pKa = 9.78DD226 pKa = 3.26TAIQRR231 pKa = 11.84PAALAYY237 pKa = 10.3SSQLRR242 pKa = 11.84TALHH246 pKa = 6.23EE247 pKa = 4.25ARR249 pKa = 11.84APLRR253 pKa = 11.84LPLFKK258 pKa = 10.29QQHH261 pKa = 6.9AYY263 pKa = 7.73TQLTAIVTRR272 pKa = 11.84QRR274 pKa = 11.84QLPVSLASDD283 pKa = 3.81NLSKK287 pKa = 10.97QPGKK291 pKa = 10.4SPRR294 pKa = 3.4
MM1 pKa = 7.69RR2 pKa = 11.84GKK4 pKa = 9.72HH5 pKa = 4.17VATRR9 pKa = 11.84SHH11 pKa = 5.44CHH13 pKa = 5.85NYY15 pKa = 9.83RR16 pKa = 11.84SAGQTQDD23 pKa = 3.12AQHH26 pKa = 6.63PLQRR30 pKa = 11.84RR31 pKa = 11.84AACCVSEE38 pKa = 4.56HH39 pKa = 6.15AWTVHH44 pKa = 5.06GASTRR49 pKa = 11.84TAASSACPGKK59 pKa = 8.38TLPIVHH65 pKa = 7.17AEE67 pKa = 3.79PTRR70 pKa = 11.84APRR73 pKa = 11.84CCHH76 pKa = 7.22LLCCPDD82 pKa = 3.34ASGMRR87 pKa = 11.84NADD90 pKa = 3.53TQRR93 pKa = 11.84NDD95 pKa = 3.25HH96 pKa = 6.74AVDD99 pKa = 3.48GCIFQKK105 pKa = 7.89EE106 pKa = 4.26TRR108 pKa = 11.84QPSNASAPSLAHH120 pKa = 5.75IVMVTLHH127 pKa = 6.89RR128 pKa = 11.84SCATQQTCRR137 pKa = 11.84SMRR140 pKa = 11.84APLPGHH146 pKa = 6.8LPSSGAHH153 pKa = 4.07SVQRR157 pKa = 11.84RR158 pKa = 11.84EE159 pKa = 3.78TRR161 pKa = 11.84AVAMHH166 pKa = 5.65STISTQHH173 pKa = 6.27KK174 pKa = 10.38GEE176 pKa = 4.26NNTKK180 pKa = 9.58KK181 pKa = 10.62ARR183 pKa = 11.84VRR185 pKa = 11.84HH186 pKa = 5.76LPASQSTVGQPTHH199 pKa = 5.87TRR201 pKa = 11.84TITQMCPAVTVGHH214 pKa = 6.92HH215 pKa = 6.79APPAPPPSTYY225 pKa = 9.78DD226 pKa = 3.26TAIQRR231 pKa = 11.84PAALAYY237 pKa = 10.3SSQLRR242 pKa = 11.84TALHH246 pKa = 6.23EE247 pKa = 4.25ARR249 pKa = 11.84APLRR253 pKa = 11.84LPLFKK258 pKa = 10.29QQHH261 pKa = 6.9AYY263 pKa = 7.73TQLTAIVTRR272 pKa = 11.84QRR274 pKa = 11.84QLPVSLASDD283 pKa = 3.81NLSKK287 pKa = 10.97QPGKK291 pKa = 10.4SPRR294 pKa = 3.4
Molecular weight: 31.96 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1476553 |
8 |
4061 |
390.8 |
42.63 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.307 ± 0.092 | 2.516 ± 0.024 |
4.517 ± 0.026 | 6.407 ± 0.041 |
2.592 ± 0.026 | 6.928 ± 0.034 |
2.723 ± 0.023 | 3.04 ± 0.023 |
5.456 ± 0.043 | 8.357 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.985 ± 0.014 | 3.629 ± 0.026 |
4.004 ± 0.043 | 3.976 ± 0.03 |
7.899 ± 0.055 | 7.426 ± 0.043 |
6.856 ± 0.049 | 6.638 ± 0.044 |
1.269 ± 0.013 | 1.473 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
