
Aspergillus fumigatus chrysovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Chrymotiviricetes; Ghabrivirales; Chrysoviridae; Alphachrysovirus
Average proteome isoelectric point is 6.36
Get precalculated fractions of proteins
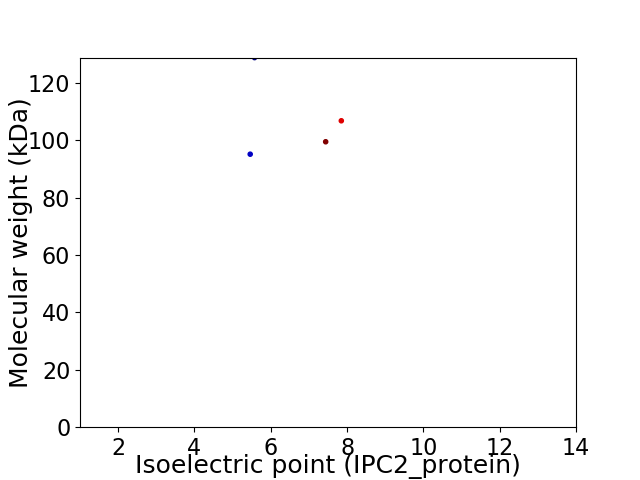
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
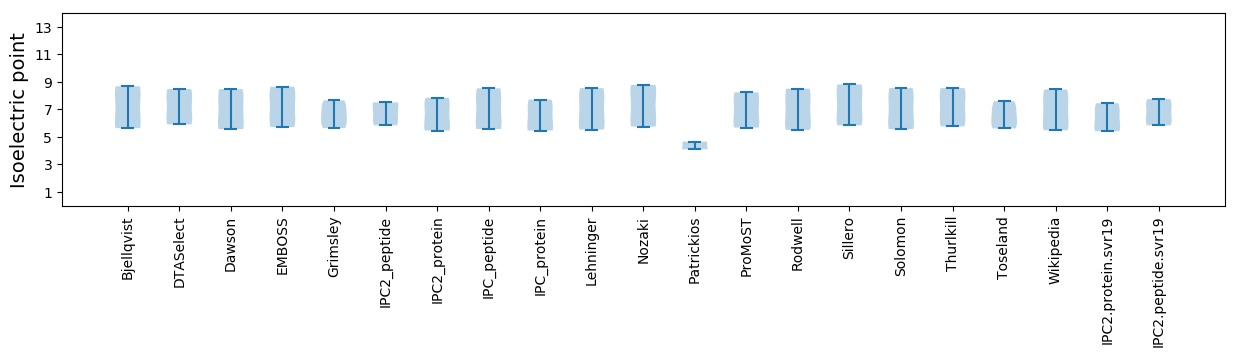
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D8L7M2|D8L7M2_9VIRU Uncharacterized protein OS=Aspergillus fumigatus chrysovirus OX=607716 PE=4 SV=1
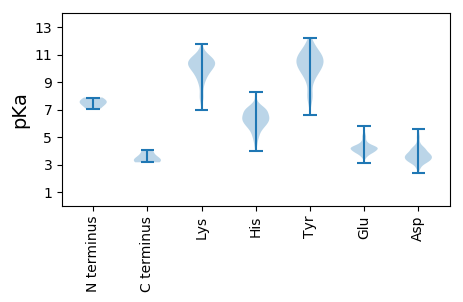
MM1 pKa = 7.07SAEE4 pKa = 4.09LRR6 pKa = 11.84VVDD9 pKa = 4.36YY10 pKa = 10.25MRR12 pKa = 11.84GLPQSYY18 pKa = 6.63QHH20 pKa = 6.97RR21 pKa = 11.84EE22 pKa = 3.39MYY24 pKa = 8.34YY25 pKa = 7.44TQFMEE30 pKa = 5.24RR31 pKa = 11.84YY32 pKa = 9.57NSTQLQEE39 pKa = 3.92VDD41 pKa = 3.36GVEE44 pKa = 4.16VEE46 pKa = 4.49KK47 pKa = 11.27GLVSDD52 pKa = 4.6GEE54 pKa = 4.3KK55 pKa = 10.56EE56 pKa = 4.24IIRR59 pKa = 11.84HH60 pKa = 5.96LNVQDD65 pKa = 3.55KK66 pKa = 8.61FTMSGFRR73 pKa = 11.84RR74 pKa = 11.84GFRR77 pKa = 11.84LAHH80 pKa = 6.31MISLGQSRR88 pKa = 11.84SVKK91 pKa = 9.38RR92 pKa = 11.84SKK94 pKa = 9.47VTSLNFQGAGEE105 pKa = 4.14WTLFASSARR114 pKa = 11.84ALYY117 pKa = 10.11SSKK120 pKa = 10.61EE121 pKa = 3.84STMNLMDD128 pKa = 4.23EE129 pKa = 4.28EE130 pKa = 4.29QKK132 pKa = 9.73RR133 pKa = 11.84TIMSKK138 pKa = 10.61CPTLSGSDD146 pKa = 3.14AGGVYY151 pKa = 8.95MAAIHH156 pKa = 6.16EE157 pKa = 4.53RR158 pKa = 11.84MSTPTDD164 pKa = 3.21LRR166 pKa = 11.84YY167 pKa = 10.31LFTKK171 pKa = 9.52MVLYY175 pKa = 10.65LYY177 pKa = 9.94DD178 pKa = 3.32YY179 pKa = 9.99TLAVVSKK186 pKa = 8.95DD187 pKa = 2.7TGMSITEE194 pKa = 4.22DD195 pKa = 3.42VAFDD199 pKa = 3.68LRR201 pKa = 11.84ANLPEE206 pKa = 4.18AARR209 pKa = 11.84LATVIDD215 pKa = 3.84HH216 pKa = 7.44DD217 pKa = 4.19IVIDD221 pKa = 3.93GDD223 pKa = 4.17LFTPEE228 pKa = 4.28QLSLLCLSGLEE239 pKa = 4.36YY240 pKa = 10.39PSVWYY245 pKa = 10.31AGEE248 pKa = 3.98GNIYY252 pKa = 10.37NSCCMEE258 pKa = 4.89KK259 pKa = 10.5DD260 pKa = 3.33DD261 pKa = 5.55LVIVSSGKK269 pKa = 9.79IEE271 pKa = 3.92VDD273 pKa = 4.0QSFLWGSPDD282 pKa = 3.63RR283 pKa = 11.84MYY285 pKa = 12.1NMMWLIAQKK294 pKa = 10.7LNAIGCLTYY303 pKa = 10.98ALEE306 pKa = 4.21NMRR309 pKa = 11.84GKK311 pKa = 10.53CKK313 pKa = 9.94MMSDD317 pKa = 3.04IVAKK321 pKa = 8.08TEE323 pKa = 3.89CRR325 pKa = 11.84EE326 pKa = 4.08VNSMVPHH333 pKa = 6.77SYY335 pKa = 11.09CMSTAFGQIRR345 pKa = 11.84EE346 pKa = 4.27RR347 pKa = 11.84QVVTKK352 pKa = 9.91MPGFFSTSLSLISDD366 pKa = 4.84LLYY369 pKa = 11.15GMCFKK374 pKa = 10.9AVASCVSEE382 pKa = 3.92TLGAMGKK389 pKa = 9.37VVSSSTPTTNEE400 pKa = 4.08TINSLMRR407 pKa = 11.84DD408 pKa = 3.38YY409 pKa = 11.35GLQHH413 pKa = 6.41TEE415 pKa = 3.08SSYY418 pKa = 11.8NYY420 pKa = 8.27MLHH423 pKa = 6.27NFEE426 pKa = 5.17LFTRR430 pKa = 11.84KK431 pKa = 7.48PTRR434 pKa = 11.84WDD436 pKa = 2.72IGQHH440 pKa = 4.01MKK442 pKa = 10.58EE443 pKa = 4.21YY444 pKa = 10.47ALKK447 pKa = 10.24LAEE450 pKa = 4.27DD451 pKa = 3.63VMMGVDD457 pKa = 3.61IPIPALLLTIPALTAVNTAFGLARR481 pKa = 11.84GWYY484 pKa = 8.07GQGDD488 pKa = 4.1ILQMTKK494 pKa = 9.08EE495 pKa = 3.97EE496 pKa = 4.36RR497 pKa = 11.84LNNTDD502 pKa = 3.7ALCAIGWMCGLRR514 pKa = 11.84DD515 pKa = 3.42VRR517 pKa = 11.84PQVFRR522 pKa = 11.84NRR524 pKa = 11.84LGRR527 pKa = 11.84KK528 pKa = 7.91QLLVNSEE535 pKa = 3.86EE536 pKa = 4.21RR537 pKa = 11.84KK538 pKa = 10.35LKK540 pKa = 11.07AEE542 pKa = 3.93ATTDD546 pKa = 3.18CRR548 pKa = 11.84VRR550 pKa = 11.84DD551 pKa = 3.55VEE553 pKa = 4.23FWIDD557 pKa = 3.24EE558 pKa = 4.39SLGGRR563 pKa = 11.84VDD565 pKa = 4.29EE566 pKa = 5.52IEE568 pKa = 3.97EE569 pKa = 4.17AGNNLFRR576 pKa = 11.84TEE578 pKa = 4.42FSGTKK583 pKa = 10.0CAMVWNYY590 pKa = 10.3EE591 pKa = 3.64YY592 pKa = 11.24GMWMEE597 pKa = 4.93AKK599 pKa = 8.84VQEE602 pKa = 4.12YY603 pKa = 9.05TRR605 pKa = 11.84LKK607 pKa = 9.98RR608 pKa = 11.84ASLAGDD614 pKa = 3.96LTQRR618 pKa = 11.84EE619 pKa = 4.55RR620 pKa = 11.84ATMSKK625 pKa = 9.88IGPTPINWGPPPSHH639 pKa = 6.52NSKK642 pKa = 10.58LSASLEE648 pKa = 3.88RR649 pKa = 11.84MRR651 pKa = 11.84AISRR655 pKa = 11.84GNAIIPSKK663 pKa = 9.98EE664 pKa = 4.0PKK666 pKa = 9.39HH667 pKa = 6.38VRR669 pKa = 11.84VSSDD673 pKa = 2.66SRR675 pKa = 11.84PVVSRR680 pKa = 11.84YY681 pKa = 9.12VEE683 pKa = 4.01EE684 pKa = 5.36GEE686 pKa = 4.62EE687 pKa = 4.0KK688 pKa = 10.44DD689 pKa = 5.16DD690 pKa = 3.55YY691 pKa = 11.55VPYY694 pKa = 10.52VKK696 pKa = 10.28PQIEE700 pKa = 4.44EE701 pKa = 4.29GEE703 pKa = 4.37KK704 pKa = 9.2ITFSEE709 pKa = 4.15IDD711 pKa = 3.42VPGDD715 pKa = 3.73GSCGIHH721 pKa = 6.63AVVKK725 pKa = 10.35DD726 pKa = 3.78LSIHH730 pKa = 5.9GRR732 pKa = 11.84IAPADD737 pKa = 3.69AQKK740 pKa = 10.27ATTLFSEE747 pKa = 4.75DD748 pKa = 3.21MASKK752 pKa = 10.31RR753 pKa = 11.84FHH755 pKa = 7.29DD756 pKa = 4.18AAEE759 pKa = 4.34LAAQCQMWGMGMDD772 pKa = 5.56LIDD775 pKa = 4.18KK776 pKa = 9.5EE777 pKa = 4.68SNRR780 pKa = 11.84VIRR783 pKa = 11.84YY784 pKa = 7.95GDD786 pKa = 3.5PDD788 pKa = 3.6ADD790 pKa = 3.58YY791 pKa = 10.87RR792 pKa = 11.84VTIVRR797 pKa = 11.84DD798 pKa = 3.66GNHH801 pKa = 6.23FKK803 pKa = 10.65AARR806 pKa = 11.84IGEE809 pKa = 4.55PGHH812 pKa = 5.74EE813 pKa = 3.9MVVRR817 pKa = 11.84SVEE820 pKa = 4.33EE821 pKa = 3.84QQSPPEE827 pKa = 3.9EE828 pKa = 4.68FIASVKK834 pKa = 10.81SLGSLFGGSPIIQQ847 pKa = 3.3
MM1 pKa = 7.07SAEE4 pKa = 4.09LRR6 pKa = 11.84VVDD9 pKa = 4.36YY10 pKa = 10.25MRR12 pKa = 11.84GLPQSYY18 pKa = 6.63QHH20 pKa = 6.97RR21 pKa = 11.84EE22 pKa = 3.39MYY24 pKa = 8.34YY25 pKa = 7.44TQFMEE30 pKa = 5.24RR31 pKa = 11.84YY32 pKa = 9.57NSTQLQEE39 pKa = 3.92VDD41 pKa = 3.36GVEE44 pKa = 4.16VEE46 pKa = 4.49KK47 pKa = 11.27GLVSDD52 pKa = 4.6GEE54 pKa = 4.3KK55 pKa = 10.56EE56 pKa = 4.24IIRR59 pKa = 11.84HH60 pKa = 5.96LNVQDD65 pKa = 3.55KK66 pKa = 8.61FTMSGFRR73 pKa = 11.84RR74 pKa = 11.84GFRR77 pKa = 11.84LAHH80 pKa = 6.31MISLGQSRR88 pKa = 11.84SVKK91 pKa = 9.38RR92 pKa = 11.84SKK94 pKa = 9.47VTSLNFQGAGEE105 pKa = 4.14WTLFASSARR114 pKa = 11.84ALYY117 pKa = 10.11SSKK120 pKa = 10.61EE121 pKa = 3.84STMNLMDD128 pKa = 4.23EE129 pKa = 4.28EE130 pKa = 4.29QKK132 pKa = 9.73RR133 pKa = 11.84TIMSKK138 pKa = 10.61CPTLSGSDD146 pKa = 3.14AGGVYY151 pKa = 8.95MAAIHH156 pKa = 6.16EE157 pKa = 4.53RR158 pKa = 11.84MSTPTDD164 pKa = 3.21LRR166 pKa = 11.84YY167 pKa = 10.31LFTKK171 pKa = 9.52MVLYY175 pKa = 10.65LYY177 pKa = 9.94DD178 pKa = 3.32YY179 pKa = 9.99TLAVVSKK186 pKa = 8.95DD187 pKa = 2.7TGMSITEE194 pKa = 4.22DD195 pKa = 3.42VAFDD199 pKa = 3.68LRR201 pKa = 11.84ANLPEE206 pKa = 4.18AARR209 pKa = 11.84LATVIDD215 pKa = 3.84HH216 pKa = 7.44DD217 pKa = 4.19IVIDD221 pKa = 3.93GDD223 pKa = 4.17LFTPEE228 pKa = 4.28QLSLLCLSGLEE239 pKa = 4.36YY240 pKa = 10.39PSVWYY245 pKa = 10.31AGEE248 pKa = 3.98GNIYY252 pKa = 10.37NSCCMEE258 pKa = 4.89KK259 pKa = 10.5DD260 pKa = 3.33DD261 pKa = 5.55LVIVSSGKK269 pKa = 9.79IEE271 pKa = 3.92VDD273 pKa = 4.0QSFLWGSPDD282 pKa = 3.63RR283 pKa = 11.84MYY285 pKa = 12.1NMMWLIAQKK294 pKa = 10.7LNAIGCLTYY303 pKa = 10.98ALEE306 pKa = 4.21NMRR309 pKa = 11.84GKK311 pKa = 10.53CKK313 pKa = 9.94MMSDD317 pKa = 3.04IVAKK321 pKa = 8.08TEE323 pKa = 3.89CRR325 pKa = 11.84EE326 pKa = 4.08VNSMVPHH333 pKa = 6.77SYY335 pKa = 11.09CMSTAFGQIRR345 pKa = 11.84EE346 pKa = 4.27RR347 pKa = 11.84QVVTKK352 pKa = 9.91MPGFFSTSLSLISDD366 pKa = 4.84LLYY369 pKa = 11.15GMCFKK374 pKa = 10.9AVASCVSEE382 pKa = 3.92TLGAMGKK389 pKa = 9.37VVSSSTPTTNEE400 pKa = 4.08TINSLMRR407 pKa = 11.84DD408 pKa = 3.38YY409 pKa = 11.35GLQHH413 pKa = 6.41TEE415 pKa = 3.08SSYY418 pKa = 11.8NYY420 pKa = 8.27MLHH423 pKa = 6.27NFEE426 pKa = 5.17LFTRR430 pKa = 11.84KK431 pKa = 7.48PTRR434 pKa = 11.84WDD436 pKa = 2.72IGQHH440 pKa = 4.01MKK442 pKa = 10.58EE443 pKa = 4.21YY444 pKa = 10.47ALKK447 pKa = 10.24LAEE450 pKa = 4.27DD451 pKa = 3.63VMMGVDD457 pKa = 3.61IPIPALLLTIPALTAVNTAFGLARR481 pKa = 11.84GWYY484 pKa = 8.07GQGDD488 pKa = 4.1ILQMTKK494 pKa = 9.08EE495 pKa = 3.97EE496 pKa = 4.36RR497 pKa = 11.84LNNTDD502 pKa = 3.7ALCAIGWMCGLRR514 pKa = 11.84DD515 pKa = 3.42VRR517 pKa = 11.84PQVFRR522 pKa = 11.84NRR524 pKa = 11.84LGRR527 pKa = 11.84KK528 pKa = 7.91QLLVNSEE535 pKa = 3.86EE536 pKa = 4.21RR537 pKa = 11.84KK538 pKa = 10.35LKK540 pKa = 11.07AEE542 pKa = 3.93ATTDD546 pKa = 3.18CRR548 pKa = 11.84VRR550 pKa = 11.84DD551 pKa = 3.55VEE553 pKa = 4.23FWIDD557 pKa = 3.24EE558 pKa = 4.39SLGGRR563 pKa = 11.84VDD565 pKa = 4.29EE566 pKa = 5.52IEE568 pKa = 3.97EE569 pKa = 4.17AGNNLFRR576 pKa = 11.84TEE578 pKa = 4.42FSGTKK583 pKa = 10.0CAMVWNYY590 pKa = 10.3EE591 pKa = 3.64YY592 pKa = 11.24GMWMEE597 pKa = 4.93AKK599 pKa = 8.84VQEE602 pKa = 4.12YY603 pKa = 9.05TRR605 pKa = 11.84LKK607 pKa = 9.98RR608 pKa = 11.84ASLAGDD614 pKa = 3.96LTQRR618 pKa = 11.84EE619 pKa = 4.55RR620 pKa = 11.84ATMSKK625 pKa = 9.88IGPTPINWGPPPSHH639 pKa = 6.52NSKK642 pKa = 10.58LSASLEE648 pKa = 3.88RR649 pKa = 11.84MRR651 pKa = 11.84AISRR655 pKa = 11.84GNAIIPSKK663 pKa = 9.98EE664 pKa = 4.0PKK666 pKa = 9.39HH667 pKa = 6.38VRR669 pKa = 11.84VSSDD673 pKa = 2.66SRR675 pKa = 11.84PVVSRR680 pKa = 11.84YY681 pKa = 9.12VEE683 pKa = 4.01EE684 pKa = 5.36GEE686 pKa = 4.62EE687 pKa = 4.0KK688 pKa = 10.44DD689 pKa = 5.16DD690 pKa = 3.55YY691 pKa = 11.55VPYY694 pKa = 10.52VKK696 pKa = 10.28PQIEE700 pKa = 4.44EE701 pKa = 4.29GEE703 pKa = 4.37KK704 pKa = 9.2ITFSEE709 pKa = 4.15IDD711 pKa = 3.42VPGDD715 pKa = 3.73GSCGIHH721 pKa = 6.63AVVKK725 pKa = 10.35DD726 pKa = 3.78LSIHH730 pKa = 5.9GRR732 pKa = 11.84IAPADD737 pKa = 3.69AQKK740 pKa = 10.27ATTLFSEE747 pKa = 4.75DD748 pKa = 3.21MASKK752 pKa = 10.31RR753 pKa = 11.84FHH755 pKa = 7.29DD756 pKa = 4.18AAEE759 pKa = 4.34LAAQCQMWGMGMDD772 pKa = 5.56LIDD775 pKa = 4.18KK776 pKa = 9.5EE777 pKa = 4.68SNRR780 pKa = 11.84VIRR783 pKa = 11.84YY784 pKa = 7.95GDD786 pKa = 3.5PDD788 pKa = 3.6ADD790 pKa = 3.58YY791 pKa = 10.87RR792 pKa = 11.84VTIVRR797 pKa = 11.84DD798 pKa = 3.66GNHH801 pKa = 6.23FKK803 pKa = 10.65AARR806 pKa = 11.84IGEE809 pKa = 4.55PGHH812 pKa = 5.74EE813 pKa = 3.9MVVRR817 pKa = 11.84SVEE820 pKa = 4.33EE821 pKa = 3.84QQSPPEE827 pKa = 3.9EE828 pKa = 4.68FIASVKK834 pKa = 10.81SLGSLFGGSPIIQQ847 pKa = 3.3
Molecular weight: 95.17 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D8L7M1|D8L7M1_9VIRU Uncharacterized protein OS=Aspergillus fumigatus chrysovirus OX=607716 PE=4 SV=1
MM1 pKa = 7.87AMATYY6 pKa = 9.78KK7 pKa = 10.92GSGGGAYY14 pKa = 10.37GPMDD18 pKa = 4.35LRR20 pKa = 11.84RR21 pKa = 11.84QVTEE25 pKa = 3.53QKK27 pKa = 10.15RR28 pKa = 11.84DD29 pKa = 3.24VFRR32 pKa = 11.84EE33 pKa = 3.8LRR35 pKa = 11.84RR36 pKa = 11.84EE37 pKa = 3.83AVALRR42 pKa = 11.84KK43 pKa = 9.52AVEE46 pKa = 4.49LRR48 pKa = 11.84NSMDD52 pKa = 2.65VWDD55 pKa = 4.12PSTRR59 pKa = 11.84YY60 pKa = 8.81GAKK63 pKa = 8.99TVRR66 pKa = 11.84EE67 pKa = 3.82KK68 pKa = 10.97SDD70 pKa = 3.11RR71 pKa = 11.84FGRR74 pKa = 11.84DD75 pKa = 2.38IGSLLAYY82 pKa = 9.47FEE84 pKa = 4.97NVHH87 pKa = 6.46SPALDD92 pKa = 3.51VIARR96 pKa = 11.84DD97 pKa = 3.55RR98 pKa = 11.84FMLRR102 pKa = 11.84YY103 pKa = 9.32QIHH106 pKa = 6.54GDD108 pKa = 3.2IDD110 pKa = 3.85RR111 pKa = 11.84DD112 pKa = 3.7AVIGANTMTVTVPVHH127 pKa = 5.16WEE129 pKa = 3.85KK130 pKa = 11.48LDD132 pKa = 3.62VTVEE136 pKa = 4.36LYY138 pKa = 9.96PGPLDD143 pKa = 4.15KK144 pKa = 11.28LMSQTKK150 pKa = 10.52LEE152 pKa = 4.12GFARR156 pKa = 11.84SGIPDD161 pKa = 3.56RR162 pKa = 11.84DD163 pKa = 3.27QVAKK167 pKa = 9.05HH168 pKa = 5.67TGWNKK173 pKa = 9.92DD174 pKa = 3.3DD175 pKa = 3.6VRR177 pKa = 11.84RR178 pKa = 11.84FPDD181 pKa = 3.05DD182 pKa = 3.97RR183 pKa = 11.84MYY185 pKa = 11.52LLQLLLEE192 pKa = 4.22QMQTGQSKK200 pKa = 8.36LTRR203 pKa = 11.84LVKK206 pKa = 10.65GFLLMLEE213 pKa = 4.26VAEE216 pKa = 5.01RR217 pKa = 11.84GPTDD221 pKa = 3.84FDD223 pKa = 4.08VGNHH227 pKa = 4.74THH229 pKa = 5.22VQYY232 pKa = 11.49APAAVINSFEE242 pKa = 4.19QPDD245 pKa = 3.19RR246 pKa = 11.84CYY248 pKa = 11.22VYY250 pKa = 10.62NSKK253 pKa = 9.79PAQIAHH259 pKa = 6.28TALLLAMCRR268 pKa = 11.84AYY270 pKa = 9.85PPPNFSSHH278 pKa = 4.49ITIPADD284 pKa = 3.42APQVCLVSQGQAPNPGGAVTLTAGLIYY311 pKa = 10.53SSLVTYY317 pKa = 11.22AMDD320 pKa = 4.09TSCTGLLQQAQIIACSLQEE339 pKa = 3.54NRR341 pKa = 11.84YY342 pKa = 7.97FSKK345 pKa = 10.97VGLPQVASLYY355 pKa = 11.15DD356 pKa = 3.45LMIPAFVAHH365 pKa = 6.75SSSLQNAMLSKK376 pKa = 10.9DD377 pKa = 3.32LAKK380 pKa = 10.66SVGRR384 pKa = 11.84VHH386 pKa = 6.89QMLGFVSAKK395 pKa = 10.53DD396 pKa = 3.67VIQASTMQATQGFDD410 pKa = 2.86VSATVRR416 pKa = 11.84QYY418 pKa = 11.82LNSNATLVHH427 pKa = 5.8QMASKK432 pKa = 10.84LSGIGIFDD440 pKa = 3.91ATPKK444 pKa = 10.62LRR446 pKa = 11.84VFDD449 pKa = 4.2YY450 pKa = 10.53MDD452 pKa = 3.32TRR454 pKa = 11.84DD455 pKa = 4.31YY456 pKa = 11.76SDD458 pKa = 5.01LLNLSLFEE466 pKa = 4.21GLWLVRR472 pKa = 11.84NATSSVHH479 pKa = 5.76NGPVGFLLNGEE490 pKa = 4.44RR491 pKa = 11.84LRR493 pKa = 11.84TADD496 pKa = 3.11KK497 pKa = 10.18TGYY500 pKa = 10.09DD501 pKa = 3.51VLVEE505 pKa = 4.08EE506 pKa = 5.0LVLAGVTVEE515 pKa = 3.95HH516 pKa = 6.84HH517 pKa = 6.48KK518 pKa = 10.77MPDD521 pKa = 3.23GAFTTSWVGVLRR533 pKa = 11.84HH534 pKa = 6.45DD535 pKa = 4.57PLRR538 pKa = 11.84QSRR541 pKa = 11.84RR542 pKa = 11.84PRR544 pKa = 11.84RR545 pKa = 11.84RR546 pKa = 11.84IEE548 pKa = 3.8TTLVRR553 pKa = 11.84EE554 pKa = 4.31CDD556 pKa = 3.81YY557 pKa = 11.14KK558 pKa = 11.34PEE560 pKa = 4.16MNLKK564 pKa = 10.11AGGRR568 pKa = 11.84KK569 pKa = 7.8VRR571 pKa = 11.84KK572 pKa = 9.02AVRR575 pKa = 11.84RR576 pKa = 11.84PTRR579 pKa = 11.84EE580 pKa = 3.83VPSAVPLSQILKK592 pKa = 7.98TPSPQRR598 pKa = 11.84QRR600 pKa = 11.84SVRR603 pKa = 11.84RR604 pKa = 11.84EE605 pKa = 4.01SPPQYY610 pKa = 10.49QSDD613 pKa = 4.19FSSSPVTEE621 pKa = 4.45DD622 pKa = 3.11QSGEE626 pKa = 4.09ALSQQASEE634 pKa = 4.23EE635 pKa = 4.22DD636 pKa = 3.88LFDD639 pKa = 4.24EE640 pKa = 5.28EE641 pKa = 5.06PPLHH645 pKa = 4.98QTQVARR651 pKa = 11.84EE652 pKa = 4.09AQRR655 pKa = 11.84KK656 pKa = 7.8VGRR659 pKa = 11.84STLEE663 pKa = 4.16RR664 pKa = 11.84ILEE667 pKa = 4.19AVEE670 pKa = 5.13EE671 pKa = 4.15VDD673 pKa = 3.6KK674 pKa = 11.69DD675 pKa = 3.69KK676 pKa = 11.73AEE678 pKa = 4.15EE679 pKa = 3.77QAKK682 pKa = 9.08VNRR685 pKa = 11.84RR686 pKa = 11.84IEE688 pKa = 4.2SFILKK693 pKa = 8.95TSPQFNPRR701 pKa = 11.84EE702 pKa = 3.87VTMRR706 pKa = 11.84ICRR709 pKa = 11.84EE710 pKa = 3.63GGLSVRR716 pKa = 11.84EE717 pKa = 3.83RR718 pKa = 11.84EE719 pKa = 3.96EE720 pKa = 3.35WLEE723 pKa = 3.77AVEE726 pKa = 4.88RR727 pKa = 11.84SIGTAKK733 pKa = 10.61ASRR736 pKa = 11.84ARR738 pKa = 11.84DD739 pKa = 3.59LRR741 pKa = 11.84EE742 pKa = 3.87LADD745 pKa = 3.42MLVAHH750 pKa = 7.11GLRR753 pKa = 11.84GATRR757 pKa = 11.84RR758 pKa = 11.84LSGNHH763 pKa = 7.07KK764 pKa = 8.29YY765 pKa = 8.97TKK767 pKa = 10.0LVNSAIEE774 pKa = 4.3SNNPSLANDD783 pKa = 4.83LLTSMRR789 pKa = 11.84VQGSAWEE796 pKa = 4.29GDD798 pKa = 4.1VIAAPQKK805 pKa = 10.09GYY807 pKa = 11.11KK808 pKa = 10.27FIANSPAWSEE818 pKa = 3.83ALQDD822 pKa = 3.4QGINPDD828 pKa = 3.35VLVANRR834 pKa = 11.84PMRR837 pKa = 11.84YY838 pKa = 9.48VLADD842 pKa = 4.22FLADD846 pKa = 3.32STTFTAQAIGRR857 pKa = 11.84LGAWLDD863 pKa = 3.75GCSVNLLPHH872 pKa = 6.68RR873 pKa = 11.84LKK875 pKa = 10.93EE876 pKa = 4.12DD877 pKa = 3.62VIPAQIPPEE886 pKa = 4.22LDD888 pKa = 2.65WYY890 pKa = 10.97RR891 pKa = 11.84NPTSMEE897 pKa = 3.2ARR899 pKa = 11.84KK900 pKa = 9.41FNKK903 pKa = 9.58EE904 pKa = 3.62DD905 pKa = 3.8SRR907 pKa = 11.84WLYY910 pKa = 10.59LAARR914 pKa = 11.84LPGGVYY920 pKa = 9.54TKK922 pKa = 11.03EE923 pKa = 3.91EE924 pKa = 4.33LVPLCAEE931 pKa = 3.6FRR933 pKa = 11.84VPVEE937 pKa = 3.83LRR939 pKa = 11.84NQLRR943 pKa = 11.84VKK945 pKa = 10.42YY946 pKa = 10.2GLFNRR951 pKa = 11.84EE952 pKa = 3.59EE953 pKa = 4.04
MM1 pKa = 7.87AMATYY6 pKa = 9.78KK7 pKa = 10.92GSGGGAYY14 pKa = 10.37GPMDD18 pKa = 4.35LRR20 pKa = 11.84RR21 pKa = 11.84QVTEE25 pKa = 3.53QKK27 pKa = 10.15RR28 pKa = 11.84DD29 pKa = 3.24VFRR32 pKa = 11.84EE33 pKa = 3.8LRR35 pKa = 11.84RR36 pKa = 11.84EE37 pKa = 3.83AVALRR42 pKa = 11.84KK43 pKa = 9.52AVEE46 pKa = 4.49LRR48 pKa = 11.84NSMDD52 pKa = 2.65VWDD55 pKa = 4.12PSTRR59 pKa = 11.84YY60 pKa = 8.81GAKK63 pKa = 8.99TVRR66 pKa = 11.84EE67 pKa = 3.82KK68 pKa = 10.97SDD70 pKa = 3.11RR71 pKa = 11.84FGRR74 pKa = 11.84DD75 pKa = 2.38IGSLLAYY82 pKa = 9.47FEE84 pKa = 4.97NVHH87 pKa = 6.46SPALDD92 pKa = 3.51VIARR96 pKa = 11.84DD97 pKa = 3.55RR98 pKa = 11.84FMLRR102 pKa = 11.84YY103 pKa = 9.32QIHH106 pKa = 6.54GDD108 pKa = 3.2IDD110 pKa = 3.85RR111 pKa = 11.84DD112 pKa = 3.7AVIGANTMTVTVPVHH127 pKa = 5.16WEE129 pKa = 3.85KK130 pKa = 11.48LDD132 pKa = 3.62VTVEE136 pKa = 4.36LYY138 pKa = 9.96PGPLDD143 pKa = 4.15KK144 pKa = 11.28LMSQTKK150 pKa = 10.52LEE152 pKa = 4.12GFARR156 pKa = 11.84SGIPDD161 pKa = 3.56RR162 pKa = 11.84DD163 pKa = 3.27QVAKK167 pKa = 9.05HH168 pKa = 5.67TGWNKK173 pKa = 9.92DD174 pKa = 3.3DD175 pKa = 3.6VRR177 pKa = 11.84RR178 pKa = 11.84FPDD181 pKa = 3.05DD182 pKa = 3.97RR183 pKa = 11.84MYY185 pKa = 11.52LLQLLLEE192 pKa = 4.22QMQTGQSKK200 pKa = 8.36LTRR203 pKa = 11.84LVKK206 pKa = 10.65GFLLMLEE213 pKa = 4.26VAEE216 pKa = 5.01RR217 pKa = 11.84GPTDD221 pKa = 3.84FDD223 pKa = 4.08VGNHH227 pKa = 4.74THH229 pKa = 5.22VQYY232 pKa = 11.49APAAVINSFEE242 pKa = 4.19QPDD245 pKa = 3.19RR246 pKa = 11.84CYY248 pKa = 11.22VYY250 pKa = 10.62NSKK253 pKa = 9.79PAQIAHH259 pKa = 6.28TALLLAMCRR268 pKa = 11.84AYY270 pKa = 9.85PPPNFSSHH278 pKa = 4.49ITIPADD284 pKa = 3.42APQVCLVSQGQAPNPGGAVTLTAGLIYY311 pKa = 10.53SSLVTYY317 pKa = 11.22AMDD320 pKa = 4.09TSCTGLLQQAQIIACSLQEE339 pKa = 3.54NRR341 pKa = 11.84YY342 pKa = 7.97FSKK345 pKa = 10.97VGLPQVASLYY355 pKa = 11.15DD356 pKa = 3.45LMIPAFVAHH365 pKa = 6.75SSSLQNAMLSKK376 pKa = 10.9DD377 pKa = 3.32LAKK380 pKa = 10.66SVGRR384 pKa = 11.84VHH386 pKa = 6.89QMLGFVSAKK395 pKa = 10.53DD396 pKa = 3.67VIQASTMQATQGFDD410 pKa = 2.86VSATVRR416 pKa = 11.84QYY418 pKa = 11.82LNSNATLVHH427 pKa = 5.8QMASKK432 pKa = 10.84LSGIGIFDD440 pKa = 3.91ATPKK444 pKa = 10.62LRR446 pKa = 11.84VFDD449 pKa = 4.2YY450 pKa = 10.53MDD452 pKa = 3.32TRR454 pKa = 11.84DD455 pKa = 4.31YY456 pKa = 11.76SDD458 pKa = 5.01LLNLSLFEE466 pKa = 4.21GLWLVRR472 pKa = 11.84NATSSVHH479 pKa = 5.76NGPVGFLLNGEE490 pKa = 4.44RR491 pKa = 11.84LRR493 pKa = 11.84TADD496 pKa = 3.11KK497 pKa = 10.18TGYY500 pKa = 10.09DD501 pKa = 3.51VLVEE505 pKa = 4.08EE506 pKa = 5.0LVLAGVTVEE515 pKa = 3.95HH516 pKa = 6.84HH517 pKa = 6.48KK518 pKa = 10.77MPDD521 pKa = 3.23GAFTTSWVGVLRR533 pKa = 11.84HH534 pKa = 6.45DD535 pKa = 4.57PLRR538 pKa = 11.84QSRR541 pKa = 11.84RR542 pKa = 11.84PRR544 pKa = 11.84RR545 pKa = 11.84RR546 pKa = 11.84IEE548 pKa = 3.8TTLVRR553 pKa = 11.84EE554 pKa = 4.31CDD556 pKa = 3.81YY557 pKa = 11.14KK558 pKa = 11.34PEE560 pKa = 4.16MNLKK564 pKa = 10.11AGGRR568 pKa = 11.84KK569 pKa = 7.8VRR571 pKa = 11.84KK572 pKa = 9.02AVRR575 pKa = 11.84RR576 pKa = 11.84PTRR579 pKa = 11.84EE580 pKa = 3.83VPSAVPLSQILKK592 pKa = 7.98TPSPQRR598 pKa = 11.84QRR600 pKa = 11.84SVRR603 pKa = 11.84RR604 pKa = 11.84EE605 pKa = 4.01SPPQYY610 pKa = 10.49QSDD613 pKa = 4.19FSSSPVTEE621 pKa = 4.45DD622 pKa = 3.11QSGEE626 pKa = 4.09ALSQQASEE634 pKa = 4.23EE635 pKa = 4.22DD636 pKa = 3.88LFDD639 pKa = 4.24EE640 pKa = 5.28EE641 pKa = 5.06PPLHH645 pKa = 4.98QTQVARR651 pKa = 11.84EE652 pKa = 4.09AQRR655 pKa = 11.84KK656 pKa = 7.8VGRR659 pKa = 11.84STLEE663 pKa = 4.16RR664 pKa = 11.84ILEE667 pKa = 4.19AVEE670 pKa = 5.13EE671 pKa = 4.15VDD673 pKa = 3.6KK674 pKa = 11.69DD675 pKa = 3.69KK676 pKa = 11.73AEE678 pKa = 4.15EE679 pKa = 3.77QAKK682 pKa = 9.08VNRR685 pKa = 11.84RR686 pKa = 11.84IEE688 pKa = 4.2SFILKK693 pKa = 8.95TSPQFNPRR701 pKa = 11.84EE702 pKa = 3.87VTMRR706 pKa = 11.84ICRR709 pKa = 11.84EE710 pKa = 3.63GGLSVRR716 pKa = 11.84EE717 pKa = 3.83RR718 pKa = 11.84EE719 pKa = 3.96EE720 pKa = 3.35WLEE723 pKa = 3.77AVEE726 pKa = 4.88RR727 pKa = 11.84SIGTAKK733 pKa = 10.61ASRR736 pKa = 11.84ARR738 pKa = 11.84DD739 pKa = 3.59LRR741 pKa = 11.84EE742 pKa = 3.87LADD745 pKa = 3.42MLVAHH750 pKa = 7.11GLRR753 pKa = 11.84GATRR757 pKa = 11.84RR758 pKa = 11.84LSGNHH763 pKa = 7.07KK764 pKa = 8.29YY765 pKa = 8.97TKK767 pKa = 10.0LVNSAIEE774 pKa = 4.3SNNPSLANDD783 pKa = 4.83LLTSMRR789 pKa = 11.84VQGSAWEE796 pKa = 4.29GDD798 pKa = 4.1VIAAPQKK805 pKa = 10.09GYY807 pKa = 11.11KK808 pKa = 10.27FIANSPAWSEE818 pKa = 3.83ALQDD822 pKa = 3.4QGINPDD828 pKa = 3.35VLVANRR834 pKa = 11.84PMRR837 pKa = 11.84YY838 pKa = 9.48VLADD842 pKa = 4.22FLADD846 pKa = 3.32STTFTAQAIGRR857 pKa = 11.84LGAWLDD863 pKa = 3.75GCSVNLLPHH872 pKa = 6.68RR873 pKa = 11.84LKK875 pKa = 10.93EE876 pKa = 4.12DD877 pKa = 3.62VIPAQIPPEE886 pKa = 4.22LDD888 pKa = 2.65WYY890 pKa = 10.97RR891 pKa = 11.84NPTSMEE897 pKa = 3.2ARR899 pKa = 11.84KK900 pKa = 9.41FNKK903 pKa = 9.58EE904 pKa = 3.62DD905 pKa = 3.8SRR907 pKa = 11.84WLYY910 pKa = 10.59LAARR914 pKa = 11.84LPGGVYY920 pKa = 9.54TKK922 pKa = 11.03EE923 pKa = 3.91EE924 pKa = 4.33LVPLCAEE931 pKa = 3.6FRR933 pKa = 11.84VPVEE937 pKa = 3.83LRR939 pKa = 11.84NQLRR943 pKa = 11.84VKK945 pKa = 10.42YY946 pKa = 10.2GLFNRR951 pKa = 11.84EE952 pKa = 3.59EE953 pKa = 4.04
Molecular weight: 106.81 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3805 |
847 |
1114 |
951.3 |
107.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.07 ± 0.497 | 1.445 ± 0.149 |
5.966 ± 0.087 | 6.965 ± 0.281 |
3.311 ± 0.755 | 6.833 ± 0.563 |
2.181 ± 0.135 | 4.494 ± 0.393 |
4.731 ± 0.146 | 9.382 ± 0.725 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.233 ± 0.423 | 3.601 ± 0.242 |
3.916 ± 0.434 | 3.89 ± 0.308 |
7.543 ± 0.51 | 7.306 ± 0.44 |
4.888 ± 0.22 | 7.937 ± 0.365 |
1.813 ± 0.234 | 3.495 ± 0.315 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
