
Tai Forest hepadnavirus
Taxonomy: Viruses; Riboviria; Pararnavirae; Artverviricota; Revtraviricetes; Blubervirales; Hepadnaviridae; unclassified Hepadnaviridae
Average proteome isoelectric point is 8.89
Get precalculated fractions of proteins
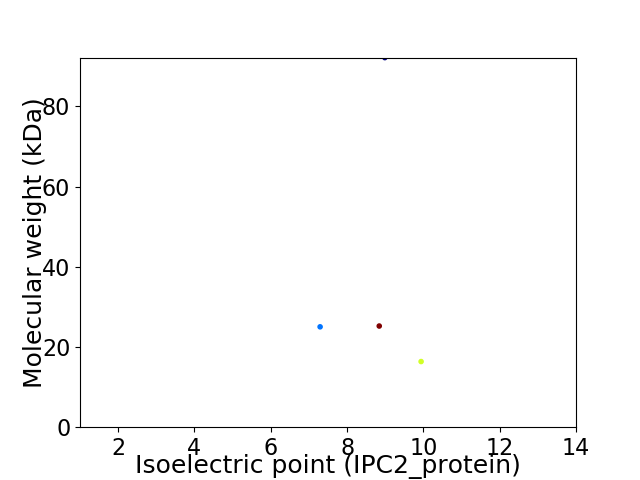
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
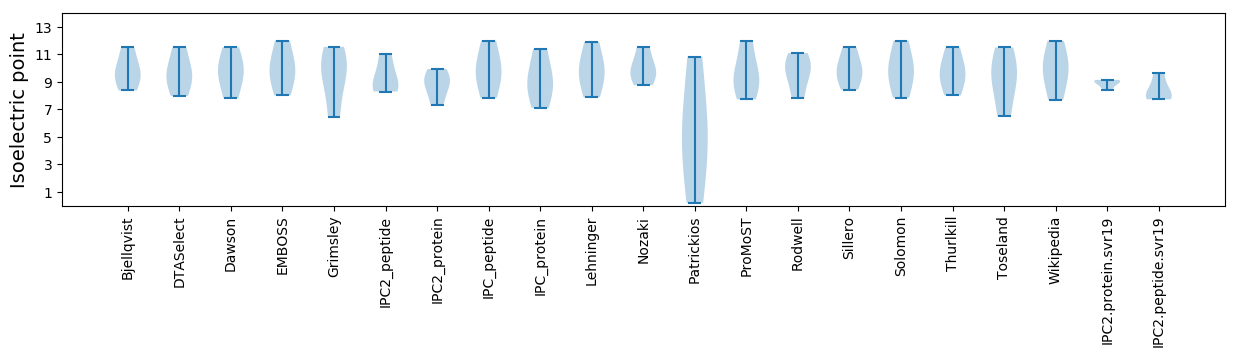
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A482KMS0|A0A482KMS0_9HEPA Core protein OS=Tai Forest hepadnavirus OX=2557875 PE=4 SV=1
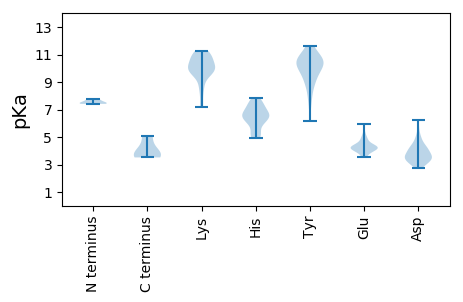
MM1 pKa = 7.44GNIASEE7 pKa = 4.2SLAPLFGLQVGYY19 pKa = 9.03FLWTKK24 pKa = 9.88ILSIAQSLDD33 pKa = 3.36SWWTSLSFPGGIPRR47 pKa = 11.84CAGLNSPFPTCDD59 pKa = 3.59HH60 pKa = 6.91SPTSCPVTCTGYY72 pKa = 10.69RR73 pKa = 11.84WMCLRR78 pKa = 11.84RR79 pKa = 11.84FIIYY83 pKa = 10.41LLVLVLCLIFLLVLLDD99 pKa = 4.11CKK101 pKa = 10.96GLIPVCPIGPSAGTRR116 pKa = 11.84GVSCRR121 pKa = 11.84TCTQPVDD128 pKa = 3.44EE129 pKa = 5.93GIWIPSCCCSKK140 pKa = 10.88SLEE143 pKa = 4.35GNCTCLAIPSSWALGRR159 pKa = 11.84YY160 pKa = 8.53LWGLASARR168 pKa = 11.84FSWLSSLQPWLQWFAGLSPIVWLLLIWMMWYY199 pKa = 9.19WGPGLLSILTPFIPLLLLFFWISVSLL225 pKa = 3.76
MM1 pKa = 7.44GNIASEE7 pKa = 4.2SLAPLFGLQVGYY19 pKa = 9.03FLWTKK24 pKa = 9.88ILSIAQSLDD33 pKa = 3.36SWWTSLSFPGGIPRR47 pKa = 11.84CAGLNSPFPTCDD59 pKa = 3.59HH60 pKa = 6.91SPTSCPVTCTGYY72 pKa = 10.69RR73 pKa = 11.84WMCLRR78 pKa = 11.84RR79 pKa = 11.84FIIYY83 pKa = 10.41LLVLVLCLIFLLVLLDD99 pKa = 4.11CKK101 pKa = 10.96GLIPVCPIGPSAGTRR116 pKa = 11.84GVSCRR121 pKa = 11.84TCTQPVDD128 pKa = 3.44EE129 pKa = 5.93GIWIPSCCCSKK140 pKa = 10.88SLEE143 pKa = 4.35GNCTCLAIPSSWALGRR159 pKa = 11.84YY160 pKa = 8.53LWGLASARR168 pKa = 11.84FSWLSSLQPWLQWFAGLSPIVWLLLIWMMWYY199 pKa = 9.19WGPGLLSILTPFIPLLLLFFWISVSLL225 pKa = 3.76
Molecular weight: 25.02 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A482KIF7|A0A482KIF7_9HEPA Surface protein OS=Tai Forest hepadnavirus OX=2557875 PE=4 SV=1
MM1 pKa = 7.74AARR4 pKa = 11.84LRR6 pKa = 11.84CRR8 pKa = 11.84LDD10 pKa = 3.05PSRR13 pKa = 11.84NILHH17 pKa = 6.72LRR19 pKa = 11.84PVGPQSSGRR28 pKa = 11.84SISRR32 pKa = 11.84SSRR35 pKa = 11.84LSPLPAAAALRR46 pKa = 11.84SFHH49 pKa = 6.84RR50 pKa = 11.84SHH52 pKa = 6.65VPLRR56 pKa = 11.84SLPSCAWSPAGPCVLRR72 pKa = 11.84FTCADD77 pKa = 3.31LRR79 pKa = 11.84RR80 pKa = 11.84CMGAPMNFARR90 pKa = 11.84WHH92 pKa = 5.19GRR94 pKa = 11.84RR95 pKa = 11.84MRR97 pKa = 11.84GLTVGTLNRR106 pKa = 11.84WQDD109 pKa = 3.73FYY111 pKa = 11.59HH112 pKa = 6.55GLFMQLWEE120 pKa = 4.2DD121 pKa = 3.66QGWSEE126 pKa = 4.4RR127 pKa = 11.84LCTYY131 pKa = 10.89VLGGCRR137 pKa = 11.84HH138 pKa = 6.08KK139 pKa = 10.88LVPTLL144 pKa = 3.79
MM1 pKa = 7.74AARR4 pKa = 11.84LRR6 pKa = 11.84CRR8 pKa = 11.84LDD10 pKa = 3.05PSRR13 pKa = 11.84NILHH17 pKa = 6.72LRR19 pKa = 11.84PVGPQSSGRR28 pKa = 11.84SISRR32 pKa = 11.84SSRR35 pKa = 11.84LSPLPAAAALRR46 pKa = 11.84SFHH49 pKa = 6.84RR50 pKa = 11.84SHH52 pKa = 6.65VPLRR56 pKa = 11.84SLPSCAWSPAGPCVLRR72 pKa = 11.84FTCADD77 pKa = 3.31LRR79 pKa = 11.84RR80 pKa = 11.84CMGAPMNFARR90 pKa = 11.84WHH92 pKa = 5.19GRR94 pKa = 11.84RR95 pKa = 11.84MRR97 pKa = 11.84GLTVGTLNRR106 pKa = 11.84WQDD109 pKa = 3.73FYY111 pKa = 11.59HH112 pKa = 6.55GLFMQLWEE120 pKa = 4.2DD121 pKa = 3.66QGWSEE126 pKa = 4.4RR127 pKa = 11.84LCTYY131 pKa = 10.89VLGGCRR137 pKa = 11.84HH138 pKa = 6.08KK139 pKa = 10.88LVPTLL144 pKa = 3.79
Molecular weight: 16.37 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1408 |
144 |
819 |
352.0 |
39.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.895 ± 0.486 | 3.338 ± 1.209 |
3.409 ± 0.658 | 2.841 ± 0.65 |
4.901 ± 0.393 | 6.463 ± 0.659 |
2.983 ± 0.682 | 3.693 ± 1.14 |
3.125 ± 1.125 | 13.636 ± 1.025 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.634 ± 0.396 | 3.196 ± 0.69 |
8.097 ± 0.269 | 2.983 ± 0.294 |
7.599 ± 1.6 | 8.949 ± 0.708 |
6.25 ± 0.771 | 5.185 ± 0.366 |
3.338 ± 0.981 | 2.486 ± 0.451 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
