
Rhodopirellula sp. SWK7
Taxonomy: cellular organisms; Bacteria; PVC group; Planctomycetes; Planctomycetia; Pirellulales; Pirellulaceae; Rhodopirellula; unclassified Rhodopirellula
Average proteome isoelectric point is 6.17
Get precalculated fractions of proteins
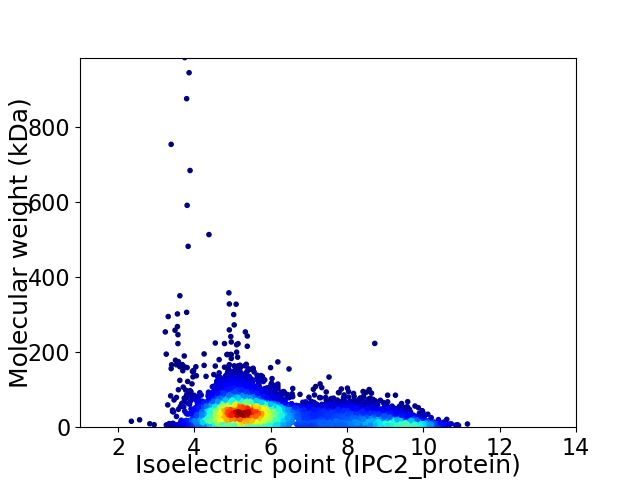
Virtual 2D-PAGE plot for 7238 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|M5TDH5|M5TDH5_9BACT DinB family protein OS=Rhodopirellula sp. SWK7 OX=595460 GN=RRSWK_05618 PE=3 SV=1
MM1 pKa = 6.54TTTVLTLDD9 pKa = 3.5SKK11 pKa = 10.88RR12 pKa = 11.84WYY14 pKa = 10.06DD15 pKa = 3.87PNSSSYY21 pKa = 9.11KK22 pKa = 7.15TTGAMKK28 pKa = 10.27VGYY31 pKa = 9.41DD32 pKa = 3.52SEE34 pKa = 4.63GSYY37 pKa = 10.23EE38 pKa = 4.2QIRR41 pKa = 11.84CSYY44 pKa = 10.77RR45 pKa = 11.84FILPIGYY52 pKa = 9.88ADD54 pKa = 4.53LFITSAVLALTVDD67 pKa = 3.79ADD69 pKa = 4.12TPNYY73 pKa = 10.67GYY75 pKa = 8.91GTDD78 pKa = 4.07TIIGPHH84 pKa = 7.04DD85 pKa = 3.58ATTAFGSIDD94 pKa = 3.84LTPYY98 pKa = 9.72ATVNLLNPKK107 pKa = 10.08GIEE110 pKa = 4.04DD111 pKa = 3.74NYY113 pKa = 10.75LGYY116 pKa = 10.93GRR118 pKa = 11.84DD119 pKa = 3.65YY120 pKa = 11.0TVTQGTISVVNVDD133 pKa = 3.56LTTAVNAAIAAGKK146 pKa = 9.36IDD148 pKa = 3.6NGGINIIWDD157 pKa = 3.75QHH159 pKa = 5.7PDD161 pKa = 2.59WYY163 pKa = 10.84GYY165 pKa = 11.0DD166 pKa = 3.51LFGDD170 pKa = 4.0NNASPTLTLTHH181 pKa = 6.24VVPPDD186 pKa = 3.58PSPYY190 pKa = 10.53GLGDD194 pKa = 3.33EE195 pKa = 4.44TVLFRR200 pKa = 11.84GATLATDD207 pKa = 4.56LGPHH211 pKa = 6.37EE212 pKa = 4.8NDD214 pKa = 3.19GTLVGGLTTVADD226 pKa = 4.19AGEE229 pKa = 3.99QGIDD233 pKa = 3.41AFDD236 pKa = 3.58MDD238 pKa = 4.51GVDD241 pKa = 3.96NKK243 pKa = 10.7LVANTFSTDD252 pKa = 3.79SEE254 pKa = 4.34WTVAAWVKK262 pKa = 10.54INSGNQKK269 pKa = 10.26GCWIKK274 pKa = 11.14SNSSASDD281 pKa = 3.35EE282 pKa = 4.4YY283 pKa = 11.68GFGLGIGNTNFDD295 pKa = 3.67TNGRR299 pKa = 11.84KK300 pKa = 9.79LIVLRR305 pKa = 11.84SGVSWHH311 pKa = 5.56VSSYY315 pKa = 10.56TFASDD320 pKa = 3.12GWHH323 pKa = 6.93HH324 pKa = 6.7IAMTRR329 pKa = 11.84SGTTNKK335 pKa = 9.82VYY337 pKa = 11.02VDD339 pKa = 3.87GVLKK343 pKa = 9.36YY344 pKa = 8.79TVAFGSNSDD353 pKa = 3.15GDD355 pKa = 4.14KK356 pKa = 10.77ISFGGFDD363 pKa = 3.64VLSSYY368 pKa = 10.96YY369 pKa = 9.84RR370 pKa = 11.84YY371 pKa = 10.17CSNRR375 pKa = 11.84MDD377 pKa = 4.34DD378 pKa = 3.8CCQFSRR384 pKa = 11.84VLSQYY389 pKa = 10.3EE390 pKa = 4.06IEE392 pKa = 4.38TLASRR397 pKa = 11.84RR398 pKa = 11.84NYY400 pKa = 10.96DD401 pKa = 3.37NTWGLNGLAHH411 pKa = 7.48RR412 pKa = 11.84YY413 pKa = 8.35CATLTDD419 pKa = 5.88DD420 pKa = 3.36VDD422 pKa = 3.86DD423 pKa = 4.3WFGTAAGAFQGSMGTEE439 pKa = 3.47ADD441 pKa = 3.68AGHH444 pKa = 7.09GGTKK448 pKa = 9.97AYY450 pKa = 10.6AFNDD454 pKa = 3.5SNDD457 pKa = 3.53YY458 pKa = 11.22VSFAEE463 pKa = 4.92HH464 pKa = 6.35YY465 pKa = 10.92SPGTSDD471 pKa = 3.94FAFSCWFRR479 pKa = 11.84LDD481 pKa = 4.5ASGTTNKK488 pKa = 9.57WIYY491 pKa = 11.06SNYY494 pKa = 9.9NGNSSIAIFTNSGKK508 pKa = 9.85IRR510 pKa = 11.84VLIRR514 pKa = 11.84DD515 pKa = 3.47EE516 pKa = 4.88SNDD519 pKa = 3.15EE520 pKa = 3.93VRR522 pKa = 11.84FDD524 pKa = 3.79STTTPAANTWYY535 pKa = 9.98HH536 pKa = 6.97LAVTWDD542 pKa = 3.65ASDD545 pKa = 3.23HH546 pKa = 4.73QARR549 pKa = 11.84LFINGTYY556 pKa = 7.39EE557 pKa = 3.77TSAINASIGNVNLTGGGVPCLGAYY581 pKa = 9.55SAGTWDD587 pKa = 3.04SNYY590 pKa = 10.02RR591 pKa = 11.84FAGGIEE597 pKa = 3.97DD598 pKa = 3.68VMIFRR603 pKa = 11.84SEE605 pKa = 3.82ISDD608 pKa = 3.43ATIASIASNPPGWDD622 pKa = 3.2GVEE625 pKa = 4.13VSAVVATIMLDD636 pKa = 3.63DD637 pKa = 3.89VTMSAAAVSVVSGEE651 pKa = 4.17SGIVLDD657 pKa = 4.82DD658 pKa = 3.58VSMAAWGVSCVSSEE672 pKa = 4.1STIVLDD678 pKa = 4.68DD679 pKa = 3.58VAMDD683 pKa = 3.74SLGTQLSVARR693 pKa = 11.84ASATITLDD701 pKa = 3.57DD702 pKa = 3.51VTMIASADD710 pKa = 4.05SIVDD714 pKa = 3.58ASSLIILDD722 pKa = 4.16DD723 pKa = 3.58VTMVAVAGAVGNASSTITLDD743 pKa = 3.61DD744 pKa = 3.42VSMITISEE752 pKa = 4.66LIVVGGASITLDD764 pKa = 3.76DD765 pKa = 4.37VSMSAAGEE773 pKa = 4.3STVTGVSTIVLDD785 pKa = 4.02NVTLSSGSQTSVSATSTIVLDD806 pKa = 4.31DD807 pKa = 4.08VEE809 pKa = 5.2LSIDD813 pKa = 3.56VPSNNRR819 pKa = 11.84ISVVLDD825 pKa = 3.62DD826 pKa = 3.96VTMASIGANIVGADD840 pKa = 3.51ATIVLDD846 pKa = 4.91DD847 pKa = 3.72ISVTVVAAVSSLTGDD862 pKa = 3.72DD863 pKa = 3.66VLFPNDD869 pKa = 4.96LFPGDD874 pKa = 4.29LFTTEE879 pKa = 4.04VFVV882 pKa = 4.36
MM1 pKa = 6.54TTTVLTLDD9 pKa = 3.5SKK11 pKa = 10.88RR12 pKa = 11.84WYY14 pKa = 10.06DD15 pKa = 3.87PNSSSYY21 pKa = 9.11KK22 pKa = 7.15TTGAMKK28 pKa = 10.27VGYY31 pKa = 9.41DD32 pKa = 3.52SEE34 pKa = 4.63GSYY37 pKa = 10.23EE38 pKa = 4.2QIRR41 pKa = 11.84CSYY44 pKa = 10.77RR45 pKa = 11.84FILPIGYY52 pKa = 9.88ADD54 pKa = 4.53LFITSAVLALTVDD67 pKa = 3.79ADD69 pKa = 4.12TPNYY73 pKa = 10.67GYY75 pKa = 8.91GTDD78 pKa = 4.07TIIGPHH84 pKa = 7.04DD85 pKa = 3.58ATTAFGSIDD94 pKa = 3.84LTPYY98 pKa = 9.72ATVNLLNPKK107 pKa = 10.08GIEE110 pKa = 4.04DD111 pKa = 3.74NYY113 pKa = 10.75LGYY116 pKa = 10.93GRR118 pKa = 11.84DD119 pKa = 3.65YY120 pKa = 11.0TVTQGTISVVNVDD133 pKa = 3.56LTTAVNAAIAAGKK146 pKa = 9.36IDD148 pKa = 3.6NGGINIIWDD157 pKa = 3.75QHH159 pKa = 5.7PDD161 pKa = 2.59WYY163 pKa = 10.84GYY165 pKa = 11.0DD166 pKa = 3.51LFGDD170 pKa = 4.0NNASPTLTLTHH181 pKa = 6.24VVPPDD186 pKa = 3.58PSPYY190 pKa = 10.53GLGDD194 pKa = 3.33EE195 pKa = 4.44TVLFRR200 pKa = 11.84GATLATDD207 pKa = 4.56LGPHH211 pKa = 6.37EE212 pKa = 4.8NDD214 pKa = 3.19GTLVGGLTTVADD226 pKa = 4.19AGEE229 pKa = 3.99QGIDD233 pKa = 3.41AFDD236 pKa = 3.58MDD238 pKa = 4.51GVDD241 pKa = 3.96NKK243 pKa = 10.7LVANTFSTDD252 pKa = 3.79SEE254 pKa = 4.34WTVAAWVKK262 pKa = 10.54INSGNQKK269 pKa = 10.26GCWIKK274 pKa = 11.14SNSSASDD281 pKa = 3.35EE282 pKa = 4.4YY283 pKa = 11.68GFGLGIGNTNFDD295 pKa = 3.67TNGRR299 pKa = 11.84KK300 pKa = 9.79LIVLRR305 pKa = 11.84SGVSWHH311 pKa = 5.56VSSYY315 pKa = 10.56TFASDD320 pKa = 3.12GWHH323 pKa = 6.93HH324 pKa = 6.7IAMTRR329 pKa = 11.84SGTTNKK335 pKa = 9.82VYY337 pKa = 11.02VDD339 pKa = 3.87GVLKK343 pKa = 9.36YY344 pKa = 8.79TVAFGSNSDD353 pKa = 3.15GDD355 pKa = 4.14KK356 pKa = 10.77ISFGGFDD363 pKa = 3.64VLSSYY368 pKa = 10.96YY369 pKa = 9.84RR370 pKa = 11.84YY371 pKa = 10.17CSNRR375 pKa = 11.84MDD377 pKa = 4.34DD378 pKa = 3.8CCQFSRR384 pKa = 11.84VLSQYY389 pKa = 10.3EE390 pKa = 4.06IEE392 pKa = 4.38TLASRR397 pKa = 11.84RR398 pKa = 11.84NYY400 pKa = 10.96DD401 pKa = 3.37NTWGLNGLAHH411 pKa = 7.48RR412 pKa = 11.84YY413 pKa = 8.35CATLTDD419 pKa = 5.88DD420 pKa = 3.36VDD422 pKa = 3.86DD423 pKa = 4.3WFGTAAGAFQGSMGTEE439 pKa = 3.47ADD441 pKa = 3.68AGHH444 pKa = 7.09GGTKK448 pKa = 9.97AYY450 pKa = 10.6AFNDD454 pKa = 3.5SNDD457 pKa = 3.53YY458 pKa = 11.22VSFAEE463 pKa = 4.92HH464 pKa = 6.35YY465 pKa = 10.92SPGTSDD471 pKa = 3.94FAFSCWFRR479 pKa = 11.84LDD481 pKa = 4.5ASGTTNKK488 pKa = 9.57WIYY491 pKa = 11.06SNYY494 pKa = 9.9NGNSSIAIFTNSGKK508 pKa = 9.85IRR510 pKa = 11.84VLIRR514 pKa = 11.84DD515 pKa = 3.47EE516 pKa = 4.88SNDD519 pKa = 3.15EE520 pKa = 3.93VRR522 pKa = 11.84FDD524 pKa = 3.79STTTPAANTWYY535 pKa = 9.98HH536 pKa = 6.97LAVTWDD542 pKa = 3.65ASDD545 pKa = 3.23HH546 pKa = 4.73QARR549 pKa = 11.84LFINGTYY556 pKa = 7.39EE557 pKa = 3.77TSAINASIGNVNLTGGGVPCLGAYY581 pKa = 9.55SAGTWDD587 pKa = 3.04SNYY590 pKa = 10.02RR591 pKa = 11.84FAGGIEE597 pKa = 3.97DD598 pKa = 3.68VMIFRR603 pKa = 11.84SEE605 pKa = 3.82ISDD608 pKa = 3.43ATIASIASNPPGWDD622 pKa = 3.2GVEE625 pKa = 4.13VSAVVATIMLDD636 pKa = 3.63DD637 pKa = 3.89VTMSAAAVSVVSGEE651 pKa = 4.17SGIVLDD657 pKa = 4.82DD658 pKa = 3.58VSMAAWGVSCVSSEE672 pKa = 4.1STIVLDD678 pKa = 4.68DD679 pKa = 3.58VAMDD683 pKa = 3.74SLGTQLSVARR693 pKa = 11.84ASATITLDD701 pKa = 3.57DD702 pKa = 3.51VTMIASADD710 pKa = 4.05SIVDD714 pKa = 3.58ASSLIILDD722 pKa = 4.16DD723 pKa = 3.58VTMVAVAGAVGNASSTITLDD743 pKa = 3.61DD744 pKa = 3.42VSMITISEE752 pKa = 4.66LIVVGGASITLDD764 pKa = 3.76DD765 pKa = 4.37VSMSAAGEE773 pKa = 4.3STVTGVSTIVLDD785 pKa = 4.02NVTLSSGSQTSVSATSTIVLDD806 pKa = 4.31DD807 pKa = 4.08VEE809 pKa = 5.2LSIDD813 pKa = 3.56VPSNNRR819 pKa = 11.84ISVVLDD825 pKa = 3.62DD826 pKa = 3.96VTMASIGANIVGADD840 pKa = 3.51ATIVLDD846 pKa = 4.91DD847 pKa = 3.72ISVTVVAAVSSLTGDD862 pKa = 3.72DD863 pKa = 3.66VLFPNDD869 pKa = 4.96LFPGDD874 pKa = 4.29LFTTEE879 pKa = 4.04VFVV882 pKa = 4.36
Molecular weight: 93.33 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|M5TUN0|M5TUN0_9BACT Uncharacterized protein OS=Rhodopirellula sp. SWK7 OX=595460 GN=RRSWK_00083 PE=4 SV=1
MM1 pKa = 6.58VRR3 pKa = 11.84LWIARR8 pKa = 11.84VRR10 pKa = 11.84IGSLVGGGFLTRR22 pKa = 11.84SASVSVILSALGVRR36 pKa = 11.84LIRR39 pKa = 11.84VILTAA44 pKa = 4.64
MM1 pKa = 6.58VRR3 pKa = 11.84LWIARR8 pKa = 11.84VRR10 pKa = 11.84IGSLVGGGFLTRR22 pKa = 11.84SASVSVILSALGVRR36 pKa = 11.84LIRR39 pKa = 11.84VILTAA44 pKa = 4.64
Molecular weight: 4.68 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2568549 |
34 |
9407 |
354.9 |
39.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.431 ± 0.031 | 1.135 ± 0.015 |
6.543 ± 0.037 | 6.162 ± 0.031 |
3.698 ± 0.021 | 7.721 ± 0.042 |
2.223 ± 0.019 | 5.18 ± 0.02 |
3.38 ± 0.035 | 9.131 ± 0.032 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.347 ± 0.019 | 3.46 ± 0.027 |
5.148 ± 0.026 | 3.736 ± 0.018 |
6.683 ± 0.04 | 7.055 ± 0.03 |
5.907 ± 0.043 | 7.217 ± 0.023 |
1.485 ± 0.015 | 2.358 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
