
Tortoise microvirus 9
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.25
Get precalculated fractions of proteins
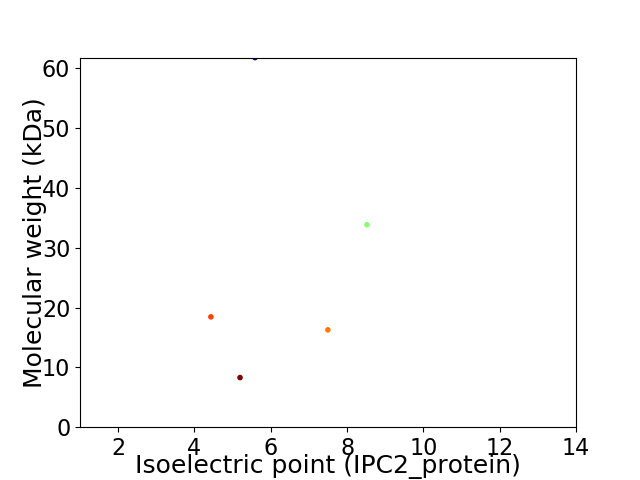
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W9A5|A0A4P8W9A5_9VIRU Uncharacterized protein OS=Tortoise microvirus 9 OX=2583198 PE=4 SV=1
MM1 pKa = 7.88KK2 pKa = 10.24YY3 pKa = 10.03RR4 pKa = 11.84RR5 pKa = 11.84PDD7 pKa = 3.41YY8 pKa = 10.9LEE10 pKa = 4.16KK11 pKa = 10.77QSFEE15 pKa = 3.91GTLYY19 pKa = 10.8SPYY22 pKa = 10.27EE23 pKa = 3.81PEE25 pKa = 3.9YY26 pKa = 10.87VLNEE30 pKa = 3.9EE31 pKa = 4.35TEE33 pKa = 4.07EE34 pKa = 4.29LEE36 pKa = 4.28IVGQINVQEE45 pKa = 4.51QIQSHH50 pKa = 6.58AGCALDD56 pKa = 5.22KK57 pKa = 10.81IFEE60 pKa = 4.44RR61 pKa = 11.84FLGDD65 pKa = 3.15IPQNNPVSTPVDD77 pKa = 3.34GVYY80 pKa = 9.38DD81 pKa = 3.69TTQRR85 pKa = 11.84ITDD88 pKa = 3.9LAEE91 pKa = 3.98LGKK94 pKa = 10.25ICDD97 pKa = 3.1IAEE100 pKa = 4.66GYY102 pKa = 9.61RR103 pKa = 11.84RR104 pKa = 11.84NNNLSAEE111 pKa = 4.14LSISQIFDD119 pKa = 4.47LISRR123 pKa = 11.84QEE125 pKa = 3.9KK126 pKa = 10.59SIAAYY131 pKa = 9.55IAAQTKK137 pKa = 10.46KK138 pKa = 10.79GDD140 pKa = 3.78NNNEE144 pKa = 3.97NEE146 pKa = 4.36KK147 pKa = 9.47KK148 pKa = 8.81TVAEE152 pKa = 4.87GEE154 pKa = 4.34QEE156 pKa = 4.3TIPTDD161 pKa = 3.77GEE163 pKa = 4.14
MM1 pKa = 7.88KK2 pKa = 10.24YY3 pKa = 10.03RR4 pKa = 11.84RR5 pKa = 11.84PDD7 pKa = 3.41YY8 pKa = 10.9LEE10 pKa = 4.16KK11 pKa = 10.77QSFEE15 pKa = 3.91GTLYY19 pKa = 10.8SPYY22 pKa = 10.27EE23 pKa = 3.81PEE25 pKa = 3.9YY26 pKa = 10.87VLNEE30 pKa = 3.9EE31 pKa = 4.35TEE33 pKa = 4.07EE34 pKa = 4.29LEE36 pKa = 4.28IVGQINVQEE45 pKa = 4.51QIQSHH50 pKa = 6.58AGCALDD56 pKa = 5.22KK57 pKa = 10.81IFEE60 pKa = 4.44RR61 pKa = 11.84FLGDD65 pKa = 3.15IPQNNPVSTPVDD77 pKa = 3.34GVYY80 pKa = 9.38DD81 pKa = 3.69TTQRR85 pKa = 11.84ITDD88 pKa = 3.9LAEE91 pKa = 3.98LGKK94 pKa = 10.25ICDD97 pKa = 3.1IAEE100 pKa = 4.66GYY102 pKa = 9.61RR103 pKa = 11.84RR104 pKa = 11.84NNNLSAEE111 pKa = 4.14LSISQIFDD119 pKa = 4.47LISRR123 pKa = 11.84QEE125 pKa = 3.9KK126 pKa = 10.59SIAAYY131 pKa = 9.55IAAQTKK137 pKa = 10.46KK138 pKa = 10.79GDD140 pKa = 3.78NNNEE144 pKa = 3.97NEE146 pKa = 4.36KK147 pKa = 9.47KK148 pKa = 8.81TVAEE152 pKa = 4.87GEE154 pKa = 4.34QEE156 pKa = 4.3TIPTDD161 pKa = 3.77GEE163 pKa = 4.14
Molecular weight: 18.44 kDa
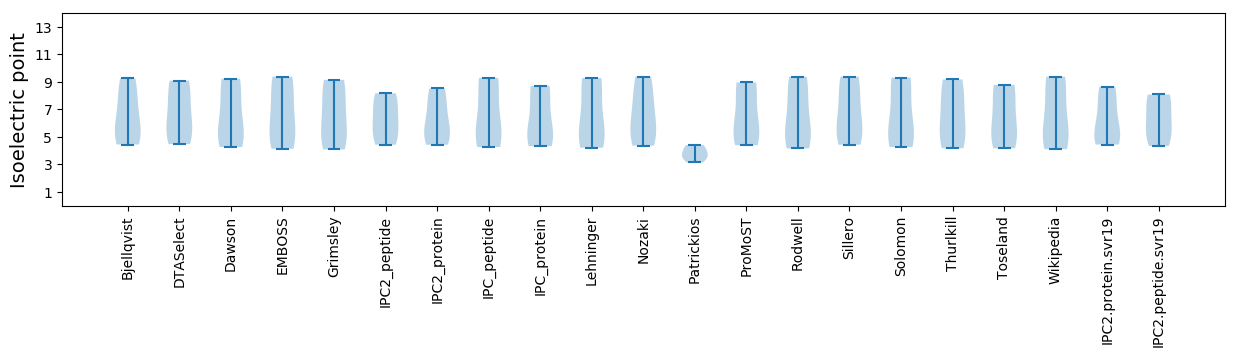
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W6S2|A0A4P8W6S2_9VIRU DNA pilot protein OS=Tortoise microvirus 9 OX=2583198 PE=4 SV=1
MM1 pKa = 6.85VVCTVVDD8 pKa = 3.7KK9 pKa = 10.57LTGHH13 pKa = 6.19IQRR16 pKa = 11.84HH17 pKa = 4.46TVFNSPMADD26 pKa = 3.53SYY28 pKa = 12.05DD29 pKa = 3.62EE30 pKa = 4.6TPTKK34 pKa = 10.78VYY36 pKa = 10.19TRR38 pKa = 11.84VNTVVACGQCLEE50 pKa = 4.61CLQQRR55 pKa = 11.84SIEE58 pKa = 3.98WSHH61 pKa = 8.0RR62 pKa = 11.84IMDD65 pKa = 3.98EE66 pKa = 4.04VSLHH70 pKa = 5.87NSCCFLTLTYY80 pKa = 10.58KK81 pKa = 10.81DD82 pKa = 3.64NPVNLVKK89 pKa = 10.25RR90 pKa = 11.84DD91 pKa = 3.34LQLFVKK97 pKa = 10.32RR98 pKa = 11.84LRR100 pKa = 11.84KK101 pKa = 9.66ALEE104 pKa = 3.96PQKK107 pKa = 10.49IRR109 pKa = 11.84YY110 pKa = 7.01FACGEE115 pKa = 4.12YY116 pKa = 10.01GAKK119 pKa = 9.7RR120 pKa = 11.84ARR122 pKa = 11.84PHH124 pKa = 4.51YY125 pKa = 10.69HH126 pKa = 5.96MIIFGWRR133 pKa = 11.84PDD135 pKa = 3.39DD136 pKa = 4.04LEE138 pKa = 4.58YY139 pKa = 11.04FFTDD143 pKa = 2.64AHH145 pKa = 6.18GFAVYY150 pKa = 9.6QSPFVAEE157 pKa = 4.26RR158 pKa = 11.84WSLGYY163 pKa = 8.8VTVGDD168 pKa = 4.02VTLEE172 pKa = 4.01TAKK175 pKa = 10.95YY176 pKa = 7.95CAKK179 pKa = 10.31YY180 pKa = 9.81LQKK183 pKa = 10.99LQDD186 pKa = 3.69TDD188 pKa = 3.71GLEE191 pKa = 4.25PPFLLMSTRR200 pKa = 11.84PGIGYY205 pKa = 9.51GVIDD209 pKa = 4.31EE210 pKa = 4.8SMLVDD215 pKa = 3.76DD216 pKa = 6.23KK217 pKa = 11.24IYY219 pKa = 11.1HH220 pKa = 5.9EE221 pKa = 4.72GNYY224 pKa = 9.65IKK226 pKa = 10.74LPRR229 pKa = 11.84YY230 pKa = 7.22YY231 pKa = 10.5CKK233 pKa = 10.58VLASRR238 pKa = 11.84CPDD241 pKa = 2.96KK242 pKa = 11.25LAVLKK247 pKa = 8.46EE248 pKa = 3.79QRR250 pKa = 11.84VRR252 pKa = 11.84KK253 pKa = 9.69AKK255 pKa = 10.4LLEE258 pKa = 4.26RR259 pKa = 11.84THH261 pKa = 6.75TEE263 pKa = 3.42LQARR267 pKa = 11.84RR268 pKa = 11.84EE269 pKa = 4.01EE270 pKa = 4.17LQARR274 pKa = 11.84LGAIRR279 pKa = 11.84CPSRR283 pKa = 11.84RR284 pKa = 11.84KK285 pKa = 9.08KK286 pKa = 11.1SKK288 pKa = 9.33MKK290 pKa = 10.43KK291 pKa = 9.52
MM1 pKa = 6.85VVCTVVDD8 pKa = 3.7KK9 pKa = 10.57LTGHH13 pKa = 6.19IQRR16 pKa = 11.84HH17 pKa = 4.46TVFNSPMADD26 pKa = 3.53SYY28 pKa = 12.05DD29 pKa = 3.62EE30 pKa = 4.6TPTKK34 pKa = 10.78VYY36 pKa = 10.19TRR38 pKa = 11.84VNTVVACGQCLEE50 pKa = 4.61CLQQRR55 pKa = 11.84SIEE58 pKa = 3.98WSHH61 pKa = 8.0RR62 pKa = 11.84IMDD65 pKa = 3.98EE66 pKa = 4.04VSLHH70 pKa = 5.87NSCCFLTLTYY80 pKa = 10.58KK81 pKa = 10.81DD82 pKa = 3.64NPVNLVKK89 pKa = 10.25RR90 pKa = 11.84DD91 pKa = 3.34LQLFVKK97 pKa = 10.32RR98 pKa = 11.84LRR100 pKa = 11.84KK101 pKa = 9.66ALEE104 pKa = 3.96PQKK107 pKa = 10.49IRR109 pKa = 11.84YY110 pKa = 7.01FACGEE115 pKa = 4.12YY116 pKa = 10.01GAKK119 pKa = 9.7RR120 pKa = 11.84ARR122 pKa = 11.84PHH124 pKa = 4.51YY125 pKa = 10.69HH126 pKa = 5.96MIIFGWRR133 pKa = 11.84PDD135 pKa = 3.39DD136 pKa = 4.04LEE138 pKa = 4.58YY139 pKa = 11.04FFTDD143 pKa = 2.64AHH145 pKa = 6.18GFAVYY150 pKa = 9.6QSPFVAEE157 pKa = 4.26RR158 pKa = 11.84WSLGYY163 pKa = 8.8VTVGDD168 pKa = 4.02VTLEE172 pKa = 4.01TAKK175 pKa = 10.95YY176 pKa = 7.95CAKK179 pKa = 10.31YY180 pKa = 9.81LQKK183 pKa = 10.99LQDD186 pKa = 3.69TDD188 pKa = 3.71GLEE191 pKa = 4.25PPFLLMSTRR200 pKa = 11.84PGIGYY205 pKa = 9.51GVIDD209 pKa = 4.31EE210 pKa = 4.8SMLVDD215 pKa = 3.76DD216 pKa = 6.23KK217 pKa = 11.24IYY219 pKa = 11.1HH220 pKa = 5.9EE221 pKa = 4.72GNYY224 pKa = 9.65IKK226 pKa = 10.74LPRR229 pKa = 11.84YY230 pKa = 7.22YY231 pKa = 10.5CKK233 pKa = 10.58VLASRR238 pKa = 11.84CPDD241 pKa = 2.96KK242 pKa = 11.25LAVLKK247 pKa = 8.46EE248 pKa = 3.79QRR250 pKa = 11.84VRR252 pKa = 11.84KK253 pKa = 9.69AKK255 pKa = 10.4LLEE258 pKa = 4.26RR259 pKa = 11.84THH261 pKa = 6.75TEE263 pKa = 3.42LQARR267 pKa = 11.84RR268 pKa = 11.84EE269 pKa = 4.01EE270 pKa = 4.17LQARR274 pKa = 11.84LGAIRR279 pKa = 11.84CPSRR283 pKa = 11.84RR284 pKa = 11.84KK285 pKa = 9.08KK286 pKa = 11.1SKK288 pKa = 9.33MKK290 pKa = 10.43KK291 pKa = 9.52
Molecular weight: 33.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1240 |
75 |
557 |
248.0 |
27.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.226 ± 1.26 | 1.452 ± 0.755 |
5.726 ± 0.087 | 5.161 ± 1.56 |
3.548 ± 0.529 | 6.855 ± 0.871 |
1.694 ± 0.418 | 4.516 ± 0.761 |
3.871 ± 1.214 | 8.065 ± 0.668 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.855 ± 0.342 | 4.435 ± 0.846 |
4.839 ± 0.635 | 4.839 ± 0.384 |
6.29 ± 0.863 | 8.145 ± 1.504 |
6.129 ± 0.125 | 7.419 ± 0.861 |
1.129 ± 0.259 | 5.806 ± 0.538 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
