
Spodoptera frugiperda ascovirus 1a (SfAV-1a)
Taxonomy: Viruses; Varidnaviria; Bamfordvirae; Nucleocytoviricota; Megaviricetes; Pimascovirales; Ascoviridae; Ascovirus
Average proteome isoelectric point is 7.14
Get precalculated fractions of proteins
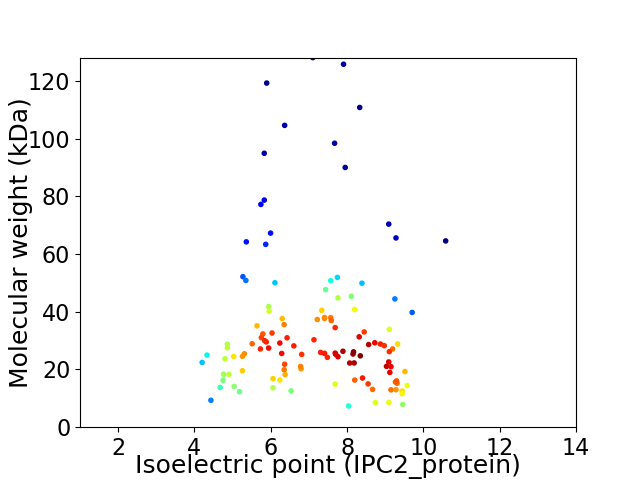
Virtual 2D-PAGE plot for 123 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q0E514|Q0E514_SFAVA 28.8 kDa Fatty acid elongase OS=Spodoptera frugiperda ascovirus 1a OX=113370 GN=ORF087 PE=4 SV=1
MM1 pKa = 7.59AMYY4 pKa = 10.33DD5 pKa = 3.16ITTSLDD11 pKa = 2.89ACLYY15 pKa = 11.04DD16 pKa = 3.74MSDD19 pKa = 3.18VSALCSRR26 pKa = 11.84LNRR29 pKa = 11.84LLSHH33 pKa = 6.91RR34 pKa = 11.84FGVVCEE40 pKa = 3.96NGKK43 pKa = 10.27VYY45 pKa = 11.09SDD47 pKa = 3.53IDD49 pKa = 3.68TLAVLVRR56 pKa = 11.84RR57 pKa = 11.84SLVPRR62 pKa = 11.84EE63 pKa = 4.28VKK65 pKa = 10.54DD66 pKa = 3.43FAYY69 pKa = 10.19GGRR72 pKa = 11.84LEE74 pKa = 4.69TYY76 pKa = 9.64NEE78 pKa = 3.52YY79 pKa = 10.8DD80 pKa = 3.66EE81 pKa = 5.8RR82 pKa = 11.84EE83 pKa = 4.03KK84 pKa = 11.23SCIRR88 pKa = 11.84NLFSRR93 pKa = 11.84CIGDD97 pKa = 4.06DD98 pKa = 3.63GSDD101 pKa = 3.46VEE103 pKa = 4.78LQRR106 pKa = 11.84NMNVDD111 pKa = 3.54SLDD114 pKa = 3.86TIIVPGLYY122 pKa = 10.15CAFFINSWYY131 pKa = 10.69RR132 pKa = 11.84FDD134 pKa = 5.33NVFDD138 pKa = 4.39RR139 pKa = 11.84NAIEE143 pKa = 4.32VNLTAARR150 pKa = 11.84TNMVNPVLITHH161 pKa = 6.48CVVCAEE167 pKa = 4.58SEE169 pKa = 4.17LDD171 pKa = 4.68PLLLWVYY178 pKa = 11.03SRR180 pKa = 11.84ACEE183 pKa = 3.84FRR185 pKa = 11.84RR186 pKa = 11.84NLFMMRR192 pKa = 11.84RR193 pKa = 11.84EE194 pKa = 4.11LVGEE198 pKa = 4.1SDD200 pKa = 4.24DD201 pKa = 4.33ADD203 pKa = 3.74SAFSDD208 pKa = 3.88SFEE211 pKa = 4.48TDD213 pKa = 3.64DD214 pKa = 4.9EE215 pKa = 6.28DD216 pKa = 5.49DD217 pKa = 3.61VV218 pKa = 4.21
MM1 pKa = 7.59AMYY4 pKa = 10.33DD5 pKa = 3.16ITTSLDD11 pKa = 2.89ACLYY15 pKa = 11.04DD16 pKa = 3.74MSDD19 pKa = 3.18VSALCSRR26 pKa = 11.84LNRR29 pKa = 11.84LLSHH33 pKa = 6.91RR34 pKa = 11.84FGVVCEE40 pKa = 3.96NGKK43 pKa = 10.27VYY45 pKa = 11.09SDD47 pKa = 3.53IDD49 pKa = 3.68TLAVLVRR56 pKa = 11.84RR57 pKa = 11.84SLVPRR62 pKa = 11.84EE63 pKa = 4.28VKK65 pKa = 10.54DD66 pKa = 3.43FAYY69 pKa = 10.19GGRR72 pKa = 11.84LEE74 pKa = 4.69TYY76 pKa = 9.64NEE78 pKa = 3.52YY79 pKa = 10.8DD80 pKa = 3.66EE81 pKa = 5.8RR82 pKa = 11.84EE83 pKa = 4.03KK84 pKa = 11.23SCIRR88 pKa = 11.84NLFSRR93 pKa = 11.84CIGDD97 pKa = 4.06DD98 pKa = 3.63GSDD101 pKa = 3.46VEE103 pKa = 4.78LQRR106 pKa = 11.84NMNVDD111 pKa = 3.54SLDD114 pKa = 3.86TIIVPGLYY122 pKa = 10.15CAFFINSWYY131 pKa = 10.69RR132 pKa = 11.84FDD134 pKa = 5.33NVFDD138 pKa = 4.39RR139 pKa = 11.84NAIEE143 pKa = 4.32VNLTAARR150 pKa = 11.84TNMVNPVLITHH161 pKa = 6.48CVVCAEE167 pKa = 4.58SEE169 pKa = 4.17LDD171 pKa = 4.68PLLLWVYY178 pKa = 11.03SRR180 pKa = 11.84ACEE183 pKa = 3.84FRR185 pKa = 11.84RR186 pKa = 11.84NLFMMRR192 pKa = 11.84RR193 pKa = 11.84EE194 pKa = 4.11LVGEE198 pKa = 4.1SDD200 pKa = 4.24DD201 pKa = 4.33ADD203 pKa = 3.74SAFSDD208 pKa = 3.88SFEE211 pKa = 4.48TDD213 pKa = 3.64DD214 pKa = 4.9EE215 pKa = 6.28DD216 pKa = 5.49DD217 pKa = 3.61VV218 pKa = 4.21
Molecular weight: 25.0 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q0E4Z0|Q0E4Z0_SFAVA 9.3 kDa OS=Spodoptera frugiperda ascovirus 1a OX=113370 GN=ORF111 PE=4 SV=1
MM1 pKa = 7.4NVTKK5 pKa = 10.89LLRR8 pKa = 11.84SYY10 pKa = 10.93NAAFTVATSTLLAPFTVMARR30 pKa = 11.84GMNVFGAEE38 pKa = 4.04SAIQFWLNVLLCCLRR53 pKa = 11.84SYY55 pKa = 11.73GMTAGIRR62 pKa = 11.84LLYY65 pKa = 10.53DD66 pKa = 3.44PAVLTSKK73 pKa = 10.69RR74 pKa = 11.84IVVVSNHH81 pKa = 4.48VNYY84 pKa = 10.66YY85 pKa = 10.33DD86 pKa = 3.82WLVLWSCMLKK96 pKa = 10.47LKK98 pKa = 10.37RR99 pKa = 11.84KK100 pKa = 9.71NVVFCAKK107 pKa = 9.67RR108 pKa = 11.84HH109 pKa = 5.12RR110 pKa = 11.84NKK112 pKa = 10.82GGFGHH117 pKa = 6.83MLNTCMKK124 pKa = 9.02ASGFIVLVTEE134 pKa = 4.06HH135 pKa = 6.07QRR137 pKa = 11.84RR138 pKa = 11.84QRR140 pKa = 11.84DD141 pKa = 3.38SGPRR145 pKa = 11.84GGEE148 pKa = 3.26IATIGRR154 pKa = 11.84VLRR157 pKa = 11.84LSISRR162 pKa = 11.84SRR164 pKa = 11.84LLNSATTSAVQLIEE178 pKa = 4.22SKK180 pKa = 10.53KK181 pKa = 9.85AARR184 pKa = 11.84STDD187 pKa = 3.06SVLRR191 pKa = 11.84ANTSSQTTPKK201 pKa = 9.1VTDD204 pKa = 3.58VPVSSTPTLLPPRR217 pKa = 11.84TRR219 pKa = 11.84GFQILMSALGEE230 pKa = 4.26SCDD233 pKa = 3.61AVVDD237 pKa = 3.93VTLSYY242 pKa = 11.06SYY244 pKa = 10.97DD245 pKa = 3.63VNVTLPKK252 pKa = 9.48SHH254 pKa = 5.84QVPRR258 pKa = 11.84SVRR261 pKa = 11.84VYY263 pKa = 9.15MRR265 pKa = 11.84KK266 pKa = 10.09LNVTFNTSEE275 pKa = 3.92TDD277 pKa = 3.96GYY279 pKa = 11.45ARR281 pKa = 11.84WLRR284 pKa = 11.84EE285 pKa = 3.51WFVQKK290 pKa = 10.87SLFIDD295 pKa = 3.88DD296 pKa = 4.52LRR298 pKa = 11.84SGFDD302 pKa = 3.17VIVPEE307 pKa = 4.01NRR309 pKa = 11.84RR310 pKa = 11.84TLTIEE315 pKa = 3.9PSPLLRR321 pKa = 11.84RR322 pKa = 11.84MYY324 pKa = 8.39TTVPSACLATTLLAMFTLVLKK345 pKa = 10.64GKK347 pKa = 10.48SITFRR352 pKa = 11.84RR353 pKa = 11.84KK354 pKa = 9.25AA355 pKa = 3.17
MM1 pKa = 7.4NVTKK5 pKa = 10.89LLRR8 pKa = 11.84SYY10 pKa = 10.93NAAFTVATSTLLAPFTVMARR30 pKa = 11.84GMNVFGAEE38 pKa = 4.04SAIQFWLNVLLCCLRR53 pKa = 11.84SYY55 pKa = 11.73GMTAGIRR62 pKa = 11.84LLYY65 pKa = 10.53DD66 pKa = 3.44PAVLTSKK73 pKa = 10.69RR74 pKa = 11.84IVVVSNHH81 pKa = 4.48VNYY84 pKa = 10.66YY85 pKa = 10.33DD86 pKa = 3.82WLVLWSCMLKK96 pKa = 10.47LKK98 pKa = 10.37RR99 pKa = 11.84KK100 pKa = 9.71NVVFCAKK107 pKa = 9.67RR108 pKa = 11.84HH109 pKa = 5.12RR110 pKa = 11.84NKK112 pKa = 10.82GGFGHH117 pKa = 6.83MLNTCMKK124 pKa = 9.02ASGFIVLVTEE134 pKa = 4.06HH135 pKa = 6.07QRR137 pKa = 11.84RR138 pKa = 11.84QRR140 pKa = 11.84DD141 pKa = 3.38SGPRR145 pKa = 11.84GGEE148 pKa = 3.26IATIGRR154 pKa = 11.84VLRR157 pKa = 11.84LSISRR162 pKa = 11.84SRR164 pKa = 11.84LLNSATTSAVQLIEE178 pKa = 4.22SKK180 pKa = 10.53KK181 pKa = 9.85AARR184 pKa = 11.84STDD187 pKa = 3.06SVLRR191 pKa = 11.84ANTSSQTTPKK201 pKa = 9.1VTDD204 pKa = 3.58VPVSSTPTLLPPRR217 pKa = 11.84TRR219 pKa = 11.84GFQILMSALGEE230 pKa = 4.26SCDD233 pKa = 3.61AVVDD237 pKa = 3.93VTLSYY242 pKa = 11.06SYY244 pKa = 10.97DD245 pKa = 3.63VNVTLPKK252 pKa = 9.48SHH254 pKa = 5.84QVPRR258 pKa = 11.84SVRR261 pKa = 11.84VYY263 pKa = 9.15MRR265 pKa = 11.84KK266 pKa = 10.09LNVTFNTSEE275 pKa = 3.92TDD277 pKa = 3.96GYY279 pKa = 11.45ARR281 pKa = 11.84WLRR284 pKa = 11.84EE285 pKa = 3.51WFVQKK290 pKa = 10.87SLFIDD295 pKa = 3.88DD296 pKa = 4.52LRR298 pKa = 11.84SGFDD302 pKa = 3.17VIVPEE307 pKa = 4.01NRR309 pKa = 11.84RR310 pKa = 11.84TLTIEE315 pKa = 3.9PSPLLRR321 pKa = 11.84RR322 pKa = 11.84MYY324 pKa = 8.39TTVPSACLATTLLAMFTLVLKK345 pKa = 10.64GKK347 pKa = 10.48SITFRR352 pKa = 11.84RR353 pKa = 11.84KK354 pKa = 9.25AA355 pKa = 3.17
Molecular weight: 39.77 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
37551 |
65 |
1157 |
305.3 |
34.59 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.255 ± 0.169 | 2.812 ± 0.128 |
6.618 ± 0.147 | 4.903 ± 0.178 |
3.417 ± 0.127 | 4.961 ± 0.17 |
2.498 ± 0.129 | 4.876 ± 0.125 |
4.495 ± 0.15 | 8.263 ± 0.189 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.807 ± 0.095 | 5.057 ± 0.14 |
3.784 ± 0.152 | 2.546 ± 0.112 |
8.271 ± 0.29 | 8.122 ± 0.304 |
6.873 ± 0.149 | 8.503 ± 0.172 |
0.956 ± 0.064 | 3.981 ± 0.157 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
