
Pacific flying fox faeces associated gemycircularvirus-6
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemykolovirus; Gemykolovirus ptero1; Pteropus associated gemykolovirus 1
Average proteome isoelectric point is 6.57
Get precalculated fractions of proteins
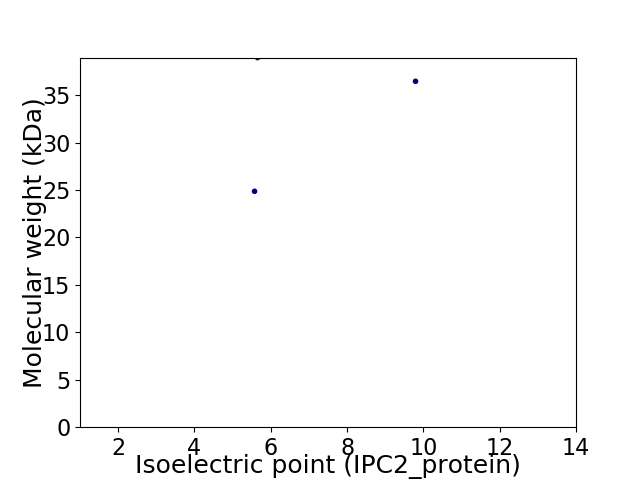
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A140CTM8|A0A140CTM8_9VIRU RepA OS=Pacific flying fox faeces associated gemycircularvirus-6 OX=1795998 PE=4 SV=1
MM1 pKa = 6.77PAKK4 pKa = 10.47YY5 pKa = 10.07KK6 pKa = 11.05LEE8 pKa = 4.3DD9 pKa = 3.33EE10 pKa = 4.52QYY12 pKa = 11.72FMLTYY17 pKa = 8.22PTIPDD22 pKa = 3.84GFDD25 pKa = 3.0TDD27 pKa = 4.31ALISVLEE34 pKa = 4.16RR35 pKa = 11.84LGCDD39 pKa = 3.23YY40 pKa = 11.01RR41 pKa = 11.84IGRR44 pKa = 11.84EE45 pKa = 3.75LHH47 pKa = 6.17QDD49 pKa = 3.92GKK51 pKa = 9.48PHH53 pKa = 5.62IHH55 pKa = 6.38VMCCFDD61 pKa = 5.54DD62 pKa = 5.09PYY64 pKa = 10.88TDD66 pKa = 3.37RR67 pKa = 11.84DD68 pKa = 3.28ARR70 pKa = 11.84ATFKK74 pKa = 10.66IGTRR78 pKa = 11.84VPNIRR83 pKa = 11.84VRR85 pKa = 11.84RR86 pKa = 11.84AKK88 pKa = 9.84PQRR91 pKa = 11.84GWDD94 pKa = 3.67YY95 pKa = 10.06VAKK98 pKa = 10.38HH99 pKa = 5.99AGQKK103 pKa = 8.71EE104 pKa = 3.79GHH106 pKa = 6.31YY107 pKa = 10.53IIGEE111 pKa = 3.99KK112 pKa = 8.88GTRR115 pKa = 11.84PGGDD119 pKa = 3.12GDD121 pKa = 3.85GSEE124 pKa = 4.88RR125 pKa = 11.84PSNDD129 pKa = 1.97AWFEE133 pKa = 4.12IILARR138 pKa = 11.84TRR140 pKa = 11.84EE141 pKa = 4.08EE142 pKa = 4.09FFDD145 pKa = 3.55AAARR149 pKa = 11.84LAPRR153 pKa = 11.84QLACNFSSLTAYY165 pKa = 10.43ADD167 pKa = 3.15WKK169 pKa = 9.31YY170 pKa = 11.08RR171 pKa = 11.84PVNVEE176 pKa = 2.96WSTPDD181 pKa = 3.34GEE183 pKa = 4.38FSVPFEE189 pKa = 4.07LRR191 pKa = 11.84DD192 pKa = 3.56WVDD195 pKa = 3.66DD196 pKa = 3.92NVRR199 pKa = 11.84GCSGMCSPFPTGGRR213 pKa = 11.84SLPRR217 pKa = 11.84EE218 pKa = 4.05
MM1 pKa = 6.77PAKK4 pKa = 10.47YY5 pKa = 10.07KK6 pKa = 11.05LEE8 pKa = 4.3DD9 pKa = 3.33EE10 pKa = 4.52QYY12 pKa = 11.72FMLTYY17 pKa = 8.22PTIPDD22 pKa = 3.84GFDD25 pKa = 3.0TDD27 pKa = 4.31ALISVLEE34 pKa = 4.16RR35 pKa = 11.84LGCDD39 pKa = 3.23YY40 pKa = 11.01RR41 pKa = 11.84IGRR44 pKa = 11.84EE45 pKa = 3.75LHH47 pKa = 6.17QDD49 pKa = 3.92GKK51 pKa = 9.48PHH53 pKa = 5.62IHH55 pKa = 6.38VMCCFDD61 pKa = 5.54DD62 pKa = 5.09PYY64 pKa = 10.88TDD66 pKa = 3.37RR67 pKa = 11.84DD68 pKa = 3.28ARR70 pKa = 11.84ATFKK74 pKa = 10.66IGTRR78 pKa = 11.84VPNIRR83 pKa = 11.84VRR85 pKa = 11.84RR86 pKa = 11.84AKK88 pKa = 9.84PQRR91 pKa = 11.84GWDD94 pKa = 3.67YY95 pKa = 10.06VAKK98 pKa = 10.38HH99 pKa = 5.99AGQKK103 pKa = 8.71EE104 pKa = 3.79GHH106 pKa = 6.31YY107 pKa = 10.53IIGEE111 pKa = 3.99KK112 pKa = 8.88GTRR115 pKa = 11.84PGGDD119 pKa = 3.12GDD121 pKa = 3.85GSEE124 pKa = 4.88RR125 pKa = 11.84PSNDD129 pKa = 1.97AWFEE133 pKa = 4.12IILARR138 pKa = 11.84TRR140 pKa = 11.84EE141 pKa = 4.08EE142 pKa = 4.09FFDD145 pKa = 3.55AAARR149 pKa = 11.84LAPRR153 pKa = 11.84QLACNFSSLTAYY165 pKa = 10.43ADD167 pKa = 3.15WKK169 pKa = 9.31YY170 pKa = 11.08RR171 pKa = 11.84PVNVEE176 pKa = 2.96WSTPDD181 pKa = 3.34GEE183 pKa = 4.38FSVPFEE189 pKa = 4.07LRR191 pKa = 11.84DD192 pKa = 3.56WVDD195 pKa = 3.66DD196 pKa = 3.92NVRR199 pKa = 11.84GCSGMCSPFPTGGRR213 pKa = 11.84SLPRR217 pKa = 11.84EE218 pKa = 4.05
Molecular weight: 24.86 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140CTM7|A0A140CTM7_9VIRU Replication-associated protein OS=Pacific flying fox faeces associated gemycircularvirus-6 OX=1795998 PE=4 SV=1
MM1 pKa = 7.87PYY3 pKa = 10.37ARR5 pKa = 11.84RR6 pKa = 11.84YY7 pKa = 3.96TTYY10 pKa = 9.75RR11 pKa = 11.84RR12 pKa = 11.84SYY14 pKa = 9.54PRR16 pKa = 11.84RR17 pKa = 11.84PYY19 pKa = 10.21QSRR22 pKa = 11.84RR23 pKa = 11.84SYY25 pKa = 10.42GVRR28 pKa = 11.84RR29 pKa = 11.84PYY31 pKa = 10.2YY32 pKa = 8.02RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84YY36 pKa = 8.34FTMPYY41 pKa = 7.56YY42 pKa = 10.33SRR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84YY48 pKa = 7.95GHH50 pKa = 6.17RR51 pKa = 11.84RR52 pKa = 11.84QYY54 pKa = 9.88GRR56 pKa = 11.84KK57 pKa = 8.13RR58 pKa = 11.84VLNITSRR65 pKa = 11.84KK66 pKa = 9.15KK67 pKa = 10.21QDD69 pKa = 2.9TMLPFTNTTASAVSGGTTFTNQAAVLVGGNTYY101 pKa = 10.06VFPWCCTARR110 pKa = 11.84DD111 pKa = 3.77NTINGSPATVFATAARR127 pKa = 11.84TATEE131 pKa = 4.25CYY133 pKa = 9.36MRR135 pKa = 11.84GLKK138 pKa = 9.86EE139 pKa = 4.23RR140 pKa = 11.84IQIQTNTGMPWQWRR154 pKa = 11.84RR155 pKa = 11.84ICFTLKK161 pKa = 10.73GDD163 pKa = 4.02VLTQYY168 pKa = 11.36NEE170 pKa = 4.01TAYY173 pKa = 9.8KK174 pKa = 10.17IYY176 pKa = 10.76DD177 pKa = 3.67EE178 pKa = 5.09TSDD181 pKa = 2.97GWSRR185 pKa = 11.84IVSNTGVASNLATAVQNLVFAGQQGKK211 pKa = 9.72DD212 pKa = 2.76WTAVFNAKK220 pKa = 9.82VDD222 pKa = 3.87DD223 pKa = 3.95RR224 pKa = 11.84RR225 pKa = 11.84LTLKK229 pKa = 10.29YY230 pKa = 10.64DD231 pKa = 3.02KK232 pKa = 10.28TVYY235 pKa = 9.75IRR237 pKa = 11.84SGNASGVMRR246 pKa = 11.84DD247 pKa = 3.24YY248 pKa = 11.31HH249 pKa = 6.47RR250 pKa = 11.84WHH252 pKa = 7.21PMNANLVYY260 pKa = 10.79DD261 pKa = 4.6DD262 pKa = 5.69DD263 pKa = 4.33EE264 pKa = 7.31AGTSMVPSYY273 pKa = 9.96FSPEE277 pKa = 3.19GRR279 pKa = 11.84AAMGDD284 pKa = 4.17YY285 pKa = 10.67YY286 pKa = 11.48VLDD289 pKa = 4.93IIAGGSGATSSDD301 pKa = 2.93QLTFNPQTTLYY312 pKa = 8.18WHH314 pKa = 6.53EE315 pKa = 4.1RR316 pKa = 3.51
MM1 pKa = 7.87PYY3 pKa = 10.37ARR5 pKa = 11.84RR6 pKa = 11.84YY7 pKa = 3.96TTYY10 pKa = 9.75RR11 pKa = 11.84RR12 pKa = 11.84SYY14 pKa = 9.54PRR16 pKa = 11.84RR17 pKa = 11.84PYY19 pKa = 10.21QSRR22 pKa = 11.84RR23 pKa = 11.84SYY25 pKa = 10.42GVRR28 pKa = 11.84RR29 pKa = 11.84PYY31 pKa = 10.2YY32 pKa = 8.02RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84YY36 pKa = 8.34FTMPYY41 pKa = 7.56YY42 pKa = 10.33SRR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84YY48 pKa = 7.95GHH50 pKa = 6.17RR51 pKa = 11.84RR52 pKa = 11.84QYY54 pKa = 9.88GRR56 pKa = 11.84KK57 pKa = 8.13RR58 pKa = 11.84VLNITSRR65 pKa = 11.84KK66 pKa = 9.15KK67 pKa = 10.21QDD69 pKa = 2.9TMLPFTNTTASAVSGGTTFTNQAAVLVGGNTYY101 pKa = 10.06VFPWCCTARR110 pKa = 11.84DD111 pKa = 3.77NTINGSPATVFATAARR127 pKa = 11.84TATEE131 pKa = 4.25CYY133 pKa = 9.36MRR135 pKa = 11.84GLKK138 pKa = 9.86EE139 pKa = 4.23RR140 pKa = 11.84IQIQTNTGMPWQWRR154 pKa = 11.84RR155 pKa = 11.84ICFTLKK161 pKa = 10.73GDD163 pKa = 4.02VLTQYY168 pKa = 11.36NEE170 pKa = 4.01TAYY173 pKa = 9.8KK174 pKa = 10.17IYY176 pKa = 10.76DD177 pKa = 3.67EE178 pKa = 5.09TSDD181 pKa = 2.97GWSRR185 pKa = 11.84IVSNTGVASNLATAVQNLVFAGQQGKK211 pKa = 9.72DD212 pKa = 2.76WTAVFNAKK220 pKa = 9.82VDD222 pKa = 3.87DD223 pKa = 3.95RR224 pKa = 11.84RR225 pKa = 11.84LTLKK229 pKa = 10.29YY230 pKa = 10.64DD231 pKa = 3.02KK232 pKa = 10.28TVYY235 pKa = 9.75IRR237 pKa = 11.84SGNASGVMRR246 pKa = 11.84DD247 pKa = 3.24YY248 pKa = 11.31HH249 pKa = 6.47RR250 pKa = 11.84WHH252 pKa = 7.21PMNANLVYY260 pKa = 10.79DD261 pKa = 4.6DD262 pKa = 5.69DD263 pKa = 4.33EE264 pKa = 7.31AGTSMVPSYY273 pKa = 9.96FSPEE277 pKa = 3.19GRR279 pKa = 11.84AAMGDD284 pKa = 4.17YY285 pKa = 10.67YY286 pKa = 11.48VLDD289 pKa = 4.93IIAGGSGATSSDD301 pKa = 2.93QLTFNPQTTLYY312 pKa = 8.18WHH314 pKa = 6.53EE315 pKa = 4.1RR316 pKa = 3.51
Molecular weight: 36.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
871 |
218 |
337 |
290.3 |
33.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.348 ± 0.265 | 2.067 ± 0.344 |
7.692 ± 1.107 | 4.937 ± 1.147 |
4.592 ± 0.618 | 8.266 ± 0.419 |
1.837 ± 0.243 | 4.133 ± 0.41 |
4.363 ± 0.607 | 5.281 ± 0.373 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.181 ± 0.281 | 3.559 ± 0.645 |
5.281 ± 0.682 | 2.87 ± 0.527 |
9.874 ± 0.683 | 5.052 ± 0.549 |
6.774 ± 1.695 | 5.396 ± 0.301 |
2.641 ± 0.278 | 5.855 ± 1.003 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
