
Lutimaribacter litoralis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Lutimaribacter
Average proteome isoelectric point is 6.22
Get precalculated fractions of proteins
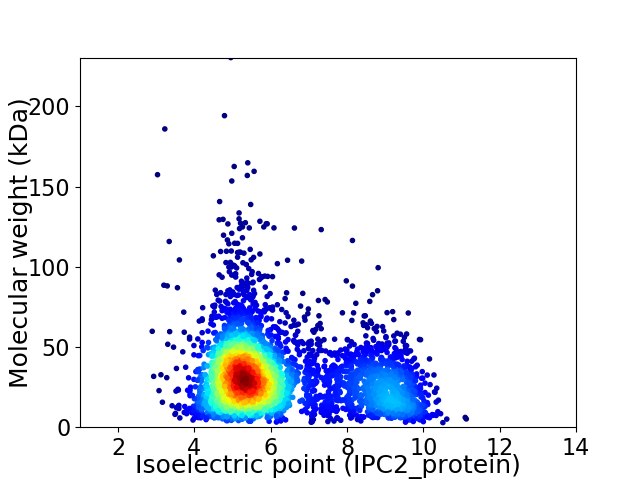
Virtual 2D-PAGE plot for 4022 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
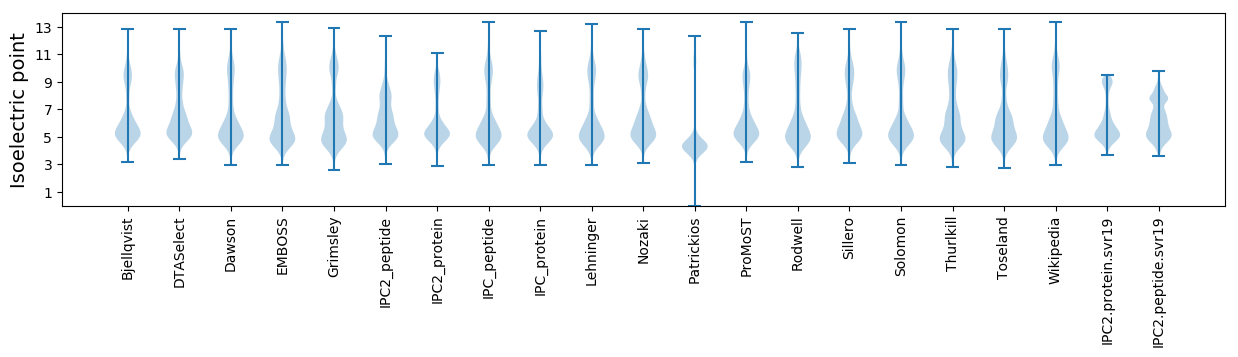
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A521B5X1|A0A521B5X1_9RHOB Glyoxylase beta-lactamase superfamily II OS=Lutimaribacter litoralis OX=1010611 GN=SAMN06265173_102155 PE=4 SV=1
MM1 pKa = 6.76PTGYY5 pKa = 10.73LVTLGNYY12 pKa = 9.53SLNSGDD18 pKa = 4.2TIGTSTYY25 pKa = 9.68SFSSASVIGSGSMSYY40 pKa = 10.68YY41 pKa = 10.07GRR43 pKa = 11.84QYY45 pKa = 9.94WYY47 pKa = 9.89QSSSYY52 pKa = 9.11RR53 pKa = 11.84TITGTYY59 pKa = 9.67YY60 pKa = 10.57EE61 pKa = 4.22ATNGNVYY68 pKa = 9.65FVPSSTLNLVQSASASTVPSYY89 pKa = 7.7TTLDD93 pKa = 3.59DD94 pKa = 3.92TVEE97 pKa = 4.17GTGADD102 pKa = 3.89DD103 pKa = 6.82LIDD106 pKa = 3.78ASYY109 pKa = 11.36GGDD112 pKa = 3.41PDD114 pKa = 4.72GDD116 pKa = 3.99HH117 pKa = 7.03VDD119 pKa = 4.34DD120 pKa = 6.46GIGTGTGGMGDD131 pKa = 3.9SIEE134 pKa = 4.21AAGGDD139 pKa = 3.66DD140 pKa = 3.25TVYY143 pKa = 10.87AGSGDD148 pKa = 3.72DD149 pKa = 4.1TITGGGGSDD158 pKa = 4.19YY159 pKa = 11.02IDD161 pKa = 4.16GGDD164 pKa = 4.01GNDD167 pKa = 3.61VIYY170 pKa = 10.99GDD172 pKa = 5.1DD173 pKa = 3.61VTSAASTAEE182 pKa = 3.93SMNWSSAGSDD192 pKa = 3.45EE193 pKa = 4.14QDD195 pKa = 2.67ISGGFTQTTGVMDD208 pKa = 3.83VNVSFSTDD216 pKa = 3.04GNPNTTFTVEE226 pKa = 4.17STTTSYY232 pKa = 11.15VGSGEE237 pKa = 4.31GFNSTSALEE246 pKa = 4.18IGSQNTGDD254 pKa = 4.07AGTVTLDD261 pKa = 3.4FTTNQPDD268 pKa = 3.69SFSDD272 pKa = 3.31NVEE275 pKa = 3.97NVSFRR280 pKa = 11.84LNDD283 pKa = 3.1VDD285 pKa = 4.5GLQDD289 pKa = 3.25GWQDD293 pKa = 3.57VITITAVDD301 pKa = 3.62ANGNTVTVLLGDD313 pKa = 3.62NGEE316 pKa = 4.59DD317 pKa = 3.37VVSGNTVTADD327 pKa = 3.64LTRR330 pKa = 11.84DD331 pKa = 3.38SYY333 pKa = 11.86TDD335 pKa = 3.37ADD337 pKa = 3.83GSVLVTIPGPVQSISIDD354 pKa = 3.62YY355 pKa = 11.23DD356 pKa = 3.25NGFANGQILIVSDD369 pKa = 3.32VHH371 pKa = 8.09FEE373 pKa = 4.4TIPLTPGDD381 pKa = 3.91DD382 pKa = 4.11TILGGAGDD390 pKa = 4.1DD391 pKa = 4.82LIYY394 pKa = 11.3GEE396 pKa = 6.22DD397 pKa = 3.77GDD399 pKa = 4.73DD400 pKa = 3.38WLGGGDD406 pKa = 4.85GSDD409 pKa = 3.35TLIGGVGNDD418 pKa = 3.79TLAGGAGGDD427 pKa = 4.14SLSSGAGMDD436 pKa = 3.57YY437 pKa = 11.07LDD439 pKa = 4.45YY440 pKa = 10.64SGSSAGVDD448 pKa = 3.42VNLGLGTASGGDD460 pKa = 3.56AEE462 pKa = 5.24GDD464 pKa = 3.78TLAGGLDD471 pKa = 4.32GIIGSDD477 pKa = 3.24WDD479 pKa = 4.03DD480 pKa = 3.49TLTGYY485 pKa = 11.13DD486 pKa = 3.49MQGIDD491 pKa = 5.86NGTAWTNVFYY501 pKa = 11.03GGGGNDD507 pKa = 4.01YY508 pKa = 10.8IDD510 pKa = 4.07GAGADD515 pKa = 3.5DD516 pKa = 4.14FLYY519 pKa = 11.07GEE521 pKa = 4.97EE522 pKa = 4.72GDD524 pKa = 3.96DD525 pKa = 4.08TILGGTGNDD534 pKa = 3.9LVDD537 pKa = 3.81GGIGNDD543 pKa = 3.58SLLGGDD549 pKa = 4.99GNDD552 pKa = 3.6TLIGGDD558 pKa = 3.84GDD560 pKa = 5.6DD561 pKa = 4.07ILSGGGGADD570 pKa = 3.05ILSGGTGNDD579 pKa = 3.33QITVGSGDD587 pKa = 3.63TATGGAGDD595 pKa = 4.39DD596 pKa = 3.73TFTIQGGGGTITIDD610 pKa = 3.18GGEE613 pKa = 4.22GGEE616 pKa = 4.39GSGDD620 pKa = 3.58TIDD623 pKa = 5.3FGGQIDD629 pKa = 4.12WGSITYY635 pKa = 9.3TNTVPSDD642 pKa = 3.42LAGSVTLADD651 pKa = 3.67GTVVTFSNIEE661 pKa = 4.06SVVICFKK668 pKa = 10.88AGTRR672 pKa = 11.84IEE674 pKa = 4.5TPHH677 pKa = 6.03GPRR680 pKa = 11.84RR681 pKa = 11.84IEE683 pKa = 4.03DD684 pKa = 4.09LRR686 pKa = 11.84PGDD689 pKa = 3.85PVITRR694 pKa = 11.84DD695 pKa = 3.55NGVQPVRR702 pKa = 11.84WAGRR706 pKa = 11.84RR707 pKa = 11.84RR708 pKa = 11.84VPGQGEE714 pKa = 3.89FAPIRR719 pKa = 11.84FAPGTVGNHH728 pKa = 5.64RR729 pKa = 11.84ALIVSPQHH737 pKa = 6.87RR738 pKa = 11.84ILHH741 pKa = 6.09HH742 pKa = 6.49SPAATLLFNSTEE754 pKa = 4.01VLVAARR760 pKa = 11.84HH761 pKa = 5.02LVNGKK766 pKa = 7.3TIRR769 pKa = 11.84QVEE772 pKa = 4.37KK773 pKa = 10.69DD774 pKa = 3.39QIDD777 pKa = 3.87YY778 pKa = 11.09VHH780 pKa = 7.08ILFDD784 pKa = 3.14RR785 pKa = 11.84HH786 pKa = 5.94EE787 pKa = 4.73IIFAEE792 pKa = 5.17GAASEE797 pKa = 4.5SFHH800 pKa = 6.95PGRR803 pKa = 11.84HH804 pKa = 5.04GLNGVMGSARR814 pKa = 11.84EE815 pKa = 3.87EE816 pKa = 4.02LFALFPEE823 pKa = 4.82LRR825 pKa = 11.84SDD827 pKa = 3.65PDD829 pKa = 3.68SFGDD833 pKa = 3.52TARR836 pKa = 11.84PCLRR840 pKa = 11.84EE841 pKa = 3.8YY842 pKa = 10.36EE843 pKa = 4.26ARR845 pKa = 11.84LLTPAA850 pKa = 4.61
MM1 pKa = 6.76PTGYY5 pKa = 10.73LVTLGNYY12 pKa = 9.53SLNSGDD18 pKa = 4.2TIGTSTYY25 pKa = 9.68SFSSASVIGSGSMSYY40 pKa = 10.68YY41 pKa = 10.07GRR43 pKa = 11.84QYY45 pKa = 9.94WYY47 pKa = 9.89QSSSYY52 pKa = 9.11RR53 pKa = 11.84TITGTYY59 pKa = 9.67YY60 pKa = 10.57EE61 pKa = 4.22ATNGNVYY68 pKa = 9.65FVPSSTLNLVQSASASTVPSYY89 pKa = 7.7TTLDD93 pKa = 3.59DD94 pKa = 3.92TVEE97 pKa = 4.17GTGADD102 pKa = 3.89DD103 pKa = 6.82LIDD106 pKa = 3.78ASYY109 pKa = 11.36GGDD112 pKa = 3.41PDD114 pKa = 4.72GDD116 pKa = 3.99HH117 pKa = 7.03VDD119 pKa = 4.34DD120 pKa = 6.46GIGTGTGGMGDD131 pKa = 3.9SIEE134 pKa = 4.21AAGGDD139 pKa = 3.66DD140 pKa = 3.25TVYY143 pKa = 10.87AGSGDD148 pKa = 3.72DD149 pKa = 4.1TITGGGGSDD158 pKa = 4.19YY159 pKa = 11.02IDD161 pKa = 4.16GGDD164 pKa = 4.01GNDD167 pKa = 3.61VIYY170 pKa = 10.99GDD172 pKa = 5.1DD173 pKa = 3.61VTSAASTAEE182 pKa = 3.93SMNWSSAGSDD192 pKa = 3.45EE193 pKa = 4.14QDD195 pKa = 2.67ISGGFTQTTGVMDD208 pKa = 3.83VNVSFSTDD216 pKa = 3.04GNPNTTFTVEE226 pKa = 4.17STTTSYY232 pKa = 11.15VGSGEE237 pKa = 4.31GFNSTSALEE246 pKa = 4.18IGSQNTGDD254 pKa = 4.07AGTVTLDD261 pKa = 3.4FTTNQPDD268 pKa = 3.69SFSDD272 pKa = 3.31NVEE275 pKa = 3.97NVSFRR280 pKa = 11.84LNDD283 pKa = 3.1VDD285 pKa = 4.5GLQDD289 pKa = 3.25GWQDD293 pKa = 3.57VITITAVDD301 pKa = 3.62ANGNTVTVLLGDD313 pKa = 3.62NGEE316 pKa = 4.59DD317 pKa = 3.37VVSGNTVTADD327 pKa = 3.64LTRR330 pKa = 11.84DD331 pKa = 3.38SYY333 pKa = 11.86TDD335 pKa = 3.37ADD337 pKa = 3.83GSVLVTIPGPVQSISIDD354 pKa = 3.62YY355 pKa = 11.23DD356 pKa = 3.25NGFANGQILIVSDD369 pKa = 3.32VHH371 pKa = 8.09FEE373 pKa = 4.4TIPLTPGDD381 pKa = 3.91DD382 pKa = 4.11TILGGAGDD390 pKa = 4.1DD391 pKa = 4.82LIYY394 pKa = 11.3GEE396 pKa = 6.22DD397 pKa = 3.77GDD399 pKa = 4.73DD400 pKa = 3.38WLGGGDD406 pKa = 4.85GSDD409 pKa = 3.35TLIGGVGNDD418 pKa = 3.79TLAGGAGGDD427 pKa = 4.14SLSSGAGMDD436 pKa = 3.57YY437 pKa = 11.07LDD439 pKa = 4.45YY440 pKa = 10.64SGSSAGVDD448 pKa = 3.42VNLGLGTASGGDD460 pKa = 3.56AEE462 pKa = 5.24GDD464 pKa = 3.78TLAGGLDD471 pKa = 4.32GIIGSDD477 pKa = 3.24WDD479 pKa = 4.03DD480 pKa = 3.49TLTGYY485 pKa = 11.13DD486 pKa = 3.49MQGIDD491 pKa = 5.86NGTAWTNVFYY501 pKa = 11.03GGGGNDD507 pKa = 4.01YY508 pKa = 10.8IDD510 pKa = 4.07GAGADD515 pKa = 3.5DD516 pKa = 4.14FLYY519 pKa = 11.07GEE521 pKa = 4.97EE522 pKa = 4.72GDD524 pKa = 3.96DD525 pKa = 4.08TILGGTGNDD534 pKa = 3.9LVDD537 pKa = 3.81GGIGNDD543 pKa = 3.58SLLGGDD549 pKa = 4.99GNDD552 pKa = 3.6TLIGGDD558 pKa = 3.84GDD560 pKa = 5.6DD561 pKa = 4.07ILSGGGGADD570 pKa = 3.05ILSGGTGNDD579 pKa = 3.33QITVGSGDD587 pKa = 3.63TATGGAGDD595 pKa = 4.39DD596 pKa = 3.73TFTIQGGGGTITIDD610 pKa = 3.18GGEE613 pKa = 4.22GGEE616 pKa = 4.39GSGDD620 pKa = 3.58TIDD623 pKa = 5.3FGGQIDD629 pKa = 4.12WGSITYY635 pKa = 9.3TNTVPSDD642 pKa = 3.42LAGSVTLADD651 pKa = 3.67GTVVTFSNIEE661 pKa = 4.06SVVICFKK668 pKa = 10.88AGTRR672 pKa = 11.84IEE674 pKa = 4.5TPHH677 pKa = 6.03GPRR680 pKa = 11.84RR681 pKa = 11.84IEE683 pKa = 4.03DD684 pKa = 4.09LRR686 pKa = 11.84PGDD689 pKa = 3.85PVITRR694 pKa = 11.84DD695 pKa = 3.55NGVQPVRR702 pKa = 11.84WAGRR706 pKa = 11.84RR707 pKa = 11.84RR708 pKa = 11.84VPGQGEE714 pKa = 3.89FAPIRR719 pKa = 11.84FAPGTVGNHH728 pKa = 5.64RR729 pKa = 11.84ALIVSPQHH737 pKa = 6.87RR738 pKa = 11.84ILHH741 pKa = 6.09HH742 pKa = 6.49SPAATLLFNSTEE754 pKa = 4.01VLVAARR760 pKa = 11.84HH761 pKa = 5.02LVNGKK766 pKa = 7.3TIRR769 pKa = 11.84QVEE772 pKa = 4.37KK773 pKa = 10.69DD774 pKa = 3.39QIDD777 pKa = 3.87YY778 pKa = 11.09VHH780 pKa = 7.08ILFDD784 pKa = 3.14RR785 pKa = 11.84HH786 pKa = 5.94EE787 pKa = 4.73IIFAEE792 pKa = 5.17GAASEE797 pKa = 4.5SFHH800 pKa = 6.95PGRR803 pKa = 11.84HH804 pKa = 5.04GLNGVMGSARR814 pKa = 11.84EE815 pKa = 3.87EE816 pKa = 4.02LFALFPEE823 pKa = 4.82LRR825 pKa = 11.84SDD827 pKa = 3.65PDD829 pKa = 3.68SFGDD833 pKa = 3.52TARR836 pKa = 11.84PCLRR840 pKa = 11.84EE841 pKa = 3.8YY842 pKa = 10.36EE843 pKa = 4.26ARR845 pKa = 11.84LLTPAA850 pKa = 4.61
Molecular weight: 86.93 kDa
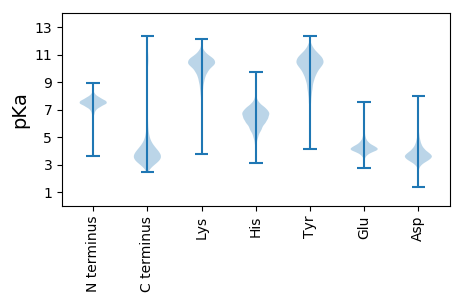
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A521E4U2|A0A521E4U2_9RHOB Acetylornithine deacetylase/Succinyl-diaminopimelate desuccinylase OS=Lutimaribacter litoralis OX=1010611 GN=SAMN06265173_11478 PE=4 SV=1
MM1 pKa = 7.61NANQLINMVLRR12 pKa = 11.84MVLRR16 pKa = 11.84KK17 pKa = 9.48AVNTGVNRR25 pKa = 11.84LAGGKK30 pKa = 9.44SGPQGVKK37 pKa = 8.45AAKK40 pKa = 9.83RR41 pKa = 11.84ARR43 pKa = 11.84QAMRR47 pKa = 11.84VTRR50 pKa = 11.84RR51 pKa = 11.84LNKK54 pKa = 9.82FF55 pKa = 3.06
MM1 pKa = 7.61NANQLINMVLRR12 pKa = 11.84MVLRR16 pKa = 11.84KK17 pKa = 9.48AVNTGVNRR25 pKa = 11.84LAGGKK30 pKa = 9.44SGPQGVKK37 pKa = 8.45AAKK40 pKa = 9.83RR41 pKa = 11.84ARR43 pKa = 11.84QAMRR47 pKa = 11.84VTRR50 pKa = 11.84RR51 pKa = 11.84LNKK54 pKa = 9.82FF55 pKa = 3.06
Molecular weight: 6.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1232936 |
25 |
2127 |
306.5 |
33.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.873 ± 0.046 | 0.895 ± 0.014 |
6.183 ± 0.037 | 5.509 ± 0.032 |
3.703 ± 0.022 | 8.665 ± 0.04 |
2.089 ± 0.018 | 5.286 ± 0.026 |
3.495 ± 0.033 | 10.042 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.924 ± 0.02 | 2.783 ± 0.022 |
5.086 ± 0.025 | 3.53 ± 0.02 |
6.392 ± 0.038 | 5.038 ± 0.026 |
5.554 ± 0.028 | 7.25 ± 0.03 |
1.378 ± 0.016 | 2.325 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
