
Sneathiella sp. DP05
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sneathiellales; Sneathiellaceae; Sneathiella
Average proteome isoelectric point is 6.03
Get precalculated fractions of proteins
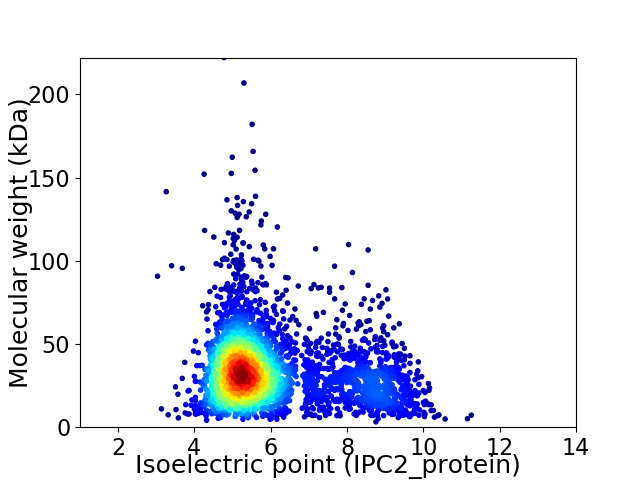
Virtual 2D-PAGE plot for 3523 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6L8W3M3|A0A6L8W3M3_9PROT ATP-binding cassette domain-containing protein OS=Sneathiella sp. DP05 OX=2606216 GN=GQE98_00695 PE=4 SV=1
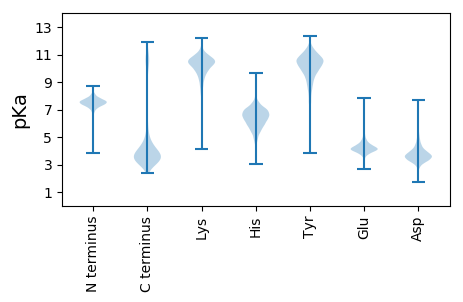
MM1 pKa = 7.26LHH3 pKa = 6.0VPLRR7 pKa = 11.84PTTLTLVFTTALPFMLFSLPVSAEE31 pKa = 3.8EE32 pKa = 3.91EE33 pKa = 4.45SFPKK37 pKa = 9.34ITGEE41 pKa = 3.98LSVEE45 pKa = 4.27VEE47 pKa = 4.29NDD49 pKa = 2.62WTYY52 pKa = 11.66QSDD55 pKa = 4.56DD56 pKa = 3.84PDD58 pKa = 5.45AEE60 pKa = 4.44INDD63 pKa = 4.59LYY65 pKa = 9.65PTVVLGTNVAFTEE78 pKa = 4.39VFSLNFEE85 pKa = 4.21ATLEE89 pKa = 4.07PVEE92 pKa = 5.17DD93 pKa = 3.87ATSDD97 pKa = 3.71RR98 pKa = 11.84AFEE101 pKa = 4.51DD102 pKa = 2.93LGGYY106 pKa = 10.26LNIITVNYY114 pKa = 10.18DD115 pKa = 3.18GEE117 pKa = 4.45AVSAYY122 pKa = 10.17AGKK125 pKa = 8.49FTPNFGIAWDD135 pKa = 3.66IAPGIFGTNLNEE147 pKa = 4.36DD148 pKa = 3.78YY149 pKa = 10.8EE150 pKa = 4.32LAEE153 pKa = 4.37MIGFGGGFHH162 pKa = 6.96FEE164 pKa = 3.94AAGEE168 pKa = 4.2HH169 pKa = 6.29TISASTFFQDD179 pKa = 4.27TSFLSNSVGTQRR191 pKa = 11.84GPLRR195 pKa = 11.84LSDD198 pKa = 3.48GGAGNTEE205 pKa = 4.14NFSSFAVALDD215 pKa = 3.71GNFQKK220 pKa = 11.09LPGFRR225 pKa = 11.84YY226 pKa = 9.91HH227 pKa = 7.4LGLSSLGAGEE237 pKa = 5.16DD238 pKa = 3.87GNDD241 pKa = 3.37RR242 pKa = 11.84QLGYY246 pKa = 10.91AIGGEE251 pKa = 4.15YY252 pKa = 10.48AFNIGEE258 pKa = 4.73DD259 pKa = 3.74VTLSPMAEE267 pKa = 3.54YY268 pKa = 10.8VYY270 pKa = 10.46FDD272 pKa = 3.61NYY274 pKa = 11.14GGIDD278 pKa = 4.03NDD280 pKa = 3.88TAKK283 pKa = 10.93YY284 pKa = 7.38FTAGLALNYY293 pKa = 8.93EE294 pKa = 4.09NWAASSTYY302 pKa = 7.81QLRR305 pKa = 11.84DD306 pKa = 3.53TEE308 pKa = 4.51VGGVMTDD315 pKa = 3.25DD316 pKa = 3.84HH317 pKa = 8.24VVDD320 pKa = 4.12LTVGYY325 pKa = 10.21VFDD328 pKa = 4.28MGLGVAAAWRR338 pKa = 11.84SAEE341 pKa = 3.93EE342 pKa = 4.35DD343 pKa = 3.81NIDD346 pKa = 4.41SEE348 pKa = 4.97GLGLLLSYY356 pKa = 10.8AIDD359 pKa = 3.82FF360 pKa = 4.48
MM1 pKa = 7.26LHH3 pKa = 6.0VPLRR7 pKa = 11.84PTTLTLVFTTALPFMLFSLPVSAEE31 pKa = 3.8EE32 pKa = 3.91EE33 pKa = 4.45SFPKK37 pKa = 9.34ITGEE41 pKa = 3.98LSVEE45 pKa = 4.27VEE47 pKa = 4.29NDD49 pKa = 2.62WTYY52 pKa = 11.66QSDD55 pKa = 4.56DD56 pKa = 3.84PDD58 pKa = 5.45AEE60 pKa = 4.44INDD63 pKa = 4.59LYY65 pKa = 9.65PTVVLGTNVAFTEE78 pKa = 4.39VFSLNFEE85 pKa = 4.21ATLEE89 pKa = 4.07PVEE92 pKa = 5.17DD93 pKa = 3.87ATSDD97 pKa = 3.71RR98 pKa = 11.84AFEE101 pKa = 4.51DD102 pKa = 2.93LGGYY106 pKa = 10.26LNIITVNYY114 pKa = 10.18DD115 pKa = 3.18GEE117 pKa = 4.45AVSAYY122 pKa = 10.17AGKK125 pKa = 8.49FTPNFGIAWDD135 pKa = 3.66IAPGIFGTNLNEE147 pKa = 4.36DD148 pKa = 3.78YY149 pKa = 10.8EE150 pKa = 4.32LAEE153 pKa = 4.37MIGFGGGFHH162 pKa = 6.96FEE164 pKa = 3.94AAGEE168 pKa = 4.2HH169 pKa = 6.29TISASTFFQDD179 pKa = 4.27TSFLSNSVGTQRR191 pKa = 11.84GPLRR195 pKa = 11.84LSDD198 pKa = 3.48GGAGNTEE205 pKa = 4.14NFSSFAVALDD215 pKa = 3.71GNFQKK220 pKa = 11.09LPGFRR225 pKa = 11.84YY226 pKa = 9.91HH227 pKa = 7.4LGLSSLGAGEE237 pKa = 5.16DD238 pKa = 3.87GNDD241 pKa = 3.37RR242 pKa = 11.84QLGYY246 pKa = 10.91AIGGEE251 pKa = 4.15YY252 pKa = 10.48AFNIGEE258 pKa = 4.73DD259 pKa = 3.74VTLSPMAEE267 pKa = 3.54YY268 pKa = 10.8VYY270 pKa = 10.46FDD272 pKa = 3.61NYY274 pKa = 11.14GGIDD278 pKa = 4.03NDD280 pKa = 3.88TAKK283 pKa = 10.93YY284 pKa = 7.38FTAGLALNYY293 pKa = 8.93EE294 pKa = 4.09NWAASSTYY302 pKa = 7.81QLRR305 pKa = 11.84DD306 pKa = 3.53TEE308 pKa = 4.51VGGVMTDD315 pKa = 3.25DD316 pKa = 3.84HH317 pKa = 8.24VVDD320 pKa = 4.12LTVGYY325 pKa = 10.21VFDD328 pKa = 4.28MGLGVAAAWRR338 pKa = 11.84SAEE341 pKa = 3.93EE342 pKa = 4.35DD343 pKa = 3.81NIDD346 pKa = 4.41SEE348 pKa = 4.97GLGLLLSYY356 pKa = 10.8AIDD359 pKa = 3.82FF360 pKa = 4.48
Molecular weight: 38.96 kDa
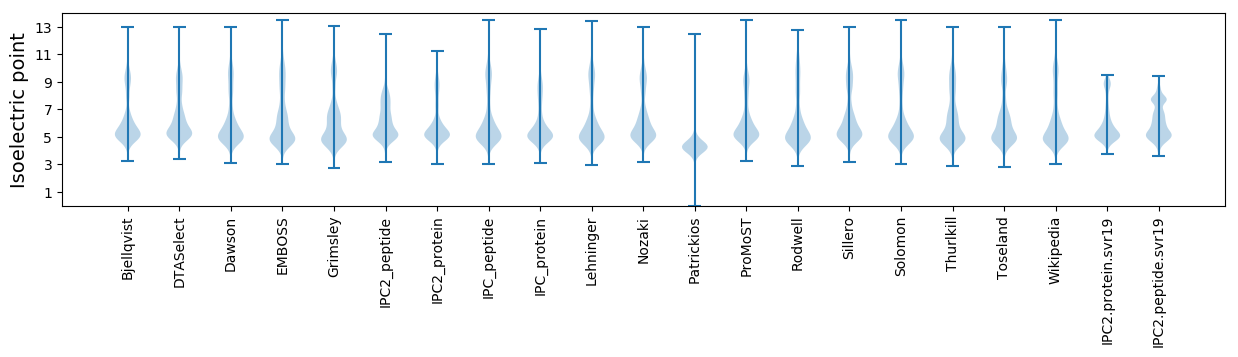
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6L8W2N0|A0A6L8W2N0_9PROT Acyl-CoA dehydrogenase OS=Sneathiella sp. DP05 OX=2606216 GN=GQE98_01630 PE=3 SV=1
MM1 pKa = 6.96TARR4 pKa = 11.84SARR7 pKa = 11.84NGRR10 pKa = 11.84NQGGKK15 pKa = 8.11SRR17 pKa = 11.84KK18 pKa = 9.35KK19 pKa = 10.16GGGGSGSGLTKK30 pKa = 10.43SSRR33 pKa = 11.84SNQQRR38 pKa = 11.84AARR41 pKa = 11.84KK42 pKa = 8.6MNLKK46 pKa = 10.04AHH48 pKa = 6.12GRR50 pKa = 11.84HH51 pKa = 5.55AARR54 pKa = 11.84RR55 pKa = 11.84QLKK58 pKa = 9.75KK59 pKa = 9.82IQAATRR65 pKa = 11.84IAAA68 pKa = 3.79
MM1 pKa = 6.96TARR4 pKa = 11.84SARR7 pKa = 11.84NGRR10 pKa = 11.84NQGGKK15 pKa = 8.11SRR17 pKa = 11.84KK18 pKa = 9.35KK19 pKa = 10.16GGGGSGSGLTKK30 pKa = 10.43SSRR33 pKa = 11.84SNQQRR38 pKa = 11.84AARR41 pKa = 11.84KK42 pKa = 8.6MNLKK46 pKa = 10.04AHH48 pKa = 6.12GRR50 pKa = 11.84HH51 pKa = 5.55AARR54 pKa = 11.84RR55 pKa = 11.84QLKK58 pKa = 9.75KK59 pKa = 9.82IQAATRR65 pKa = 11.84IAAA68 pKa = 3.79
Molecular weight: 7.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1128886 |
29 |
2065 |
320.4 |
35.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.006 ± 0.043 | 0.898 ± 0.011 |
5.742 ± 0.033 | 6.514 ± 0.039 |
4.093 ± 0.028 | 8.025 ± 0.036 |
1.965 ± 0.018 | 6.543 ± 0.031 |
4.729 ± 0.033 | 10.032 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.727 ± 0.017 | 3.468 ± 0.021 |
4.532 ± 0.022 | 3.067 ± 0.021 |
5.537 ± 0.032 | 5.946 ± 0.027 |
5.339 ± 0.024 | 7.018 ± 0.032 |
1.202 ± 0.015 | 2.62 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
