
Beilong virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; Orthoparamyxovirinae; Jeilongvirus; Beilong jeilongvirus
Average proteome isoelectric point is 6.76
Get precalculated fractions of proteins
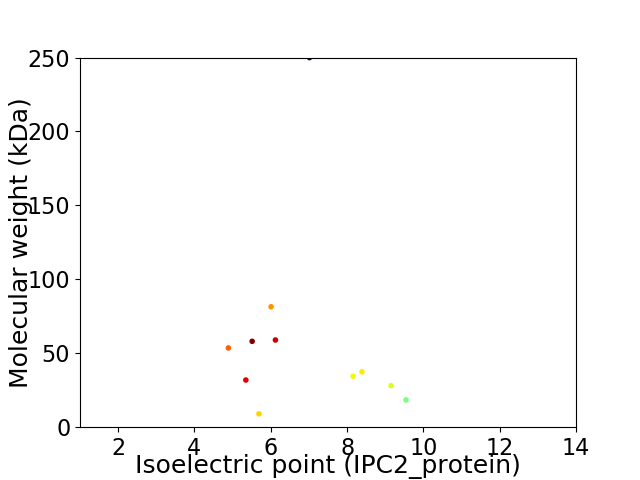
Virtual 2D-PAGE plot for 11 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q287Y1|Q287Y1_9MONO Nucleocapsid OS=Beilong virus OX=341053 GN=N PE=3 SV=1
MM1 pKa = 7.73SDD3 pKa = 3.66YY4 pKa = 11.3NPDD7 pKa = 3.21VLQQLVKK14 pKa = 10.94DD15 pKa = 4.5GIKK18 pKa = 9.87TIEE21 pKa = 4.64LLQQSPEE28 pKa = 4.27DD29 pKa = 3.66FQKK32 pKa = 10.02TYY34 pKa = 10.56GRR36 pKa = 11.84SAIQEE41 pKa = 3.9PSTRR45 pKa = 11.84ARR47 pKa = 11.84IQSWEE52 pKa = 4.05SRR54 pKa = 11.84NPDD57 pKa = 2.98HH58 pKa = 8.22DD59 pKa = 3.87YY60 pKa = 11.2TGNKK64 pKa = 7.4NQRR67 pKa = 11.84SEE69 pKa = 4.25GAKK72 pKa = 9.54EE73 pKa = 3.59RR74 pKa = 11.84ANKK77 pKa = 10.16SEE79 pKa = 4.19SAGTASADD87 pKa = 3.54GGHH90 pKa = 6.72GDD92 pKa = 3.68KK93 pKa = 10.73PSNNGGDD100 pKa = 3.65SQNEE104 pKa = 4.14YY105 pKa = 10.4QGSDD109 pKa = 3.36QQVWDD114 pKa = 3.49AAYY117 pKa = 11.07NDD119 pKa = 4.47GNSGGAWGGPTGGLPTAGEE138 pKa = 3.97RR139 pKa = 11.84GYY141 pKa = 9.98PITTGNQEE149 pKa = 4.06FEE151 pKa = 4.82GYY153 pKa = 10.34HH154 pKa = 6.45PDD156 pKa = 3.57GPVDD160 pKa = 3.27ARR162 pKa = 11.84EE163 pKa = 3.96YY164 pKa = 10.21NQISSMDD171 pKa = 3.72HH172 pKa = 5.74EE173 pKa = 4.57MSAAEE178 pKa = 4.06TLGSSTQLTTMRR190 pKa = 11.84NATTDD195 pKa = 3.38DD196 pKa = 3.37FAKK199 pKa = 10.19IFEE202 pKa = 4.48EE203 pKa = 4.86GTPKK207 pKa = 9.59VHH209 pKa = 6.94RR210 pKa = 11.84RR211 pKa = 11.84LRR213 pKa = 11.84GITAVVPAQKK223 pKa = 10.33QPGAVGGPVKK233 pKa = 10.57KK234 pKa = 9.83GTDD237 pKa = 3.22GSTASTLLGDD247 pKa = 3.97VPLSGSGAIPNVHH260 pKa = 7.08PSLLHH265 pKa = 4.98QPKK268 pKa = 10.34KK269 pKa = 10.18NAHH272 pKa = 6.42AEE274 pKa = 4.0NAQGSVQDD282 pKa = 3.7VSTTGAIGQSDD293 pKa = 3.88EE294 pKa = 4.43AAHH297 pKa = 6.78FDD299 pKa = 4.02QEE301 pKa = 4.88IEE303 pKa = 4.23GKK305 pKa = 10.62LNLVLKK311 pKa = 10.3EE312 pKa = 4.38LDD314 pKa = 4.63LISKK318 pKa = 10.15KK319 pKa = 10.38LDD321 pKa = 3.35YY322 pKa = 10.97LPEE325 pKa = 5.38IKK327 pKa = 10.58EE328 pKa = 4.09EE329 pKa = 3.65IRR331 pKa = 11.84NINKK335 pKa = 10.34KK336 pKa = 8.04ITNLSLGLSTVEE348 pKa = 4.21NYY350 pKa = 9.38IKK352 pKa = 11.05SMMIIIPSSGKK363 pKa = 9.59PDD365 pKa = 3.5VNDD368 pKa = 3.66TAEE371 pKa = 4.08VNPDD375 pKa = 3.29LKK377 pKa = 11.16AVIGRR382 pKa = 11.84DD383 pKa = 3.0KK384 pKa = 11.08TRR386 pKa = 11.84GLKK389 pKa = 9.92EE390 pKa = 4.15VTTHH394 pKa = 7.59RR395 pKa = 11.84SDD397 pKa = 5.14LEE399 pKa = 4.16SLDD402 pKa = 4.21LLSPATSEE410 pKa = 4.3IDD412 pKa = 3.66PKK414 pKa = 11.45YY415 pKa = 9.84LTQDD419 pKa = 3.09LDD421 pKa = 3.99FTKK424 pKa = 10.93SNAANFVPGNDD435 pKa = 2.96ASSFYY440 pKa = 10.54TIVAMIKK447 pKa = 10.89GEE449 pKa = 4.07VTDD452 pKa = 3.54VKK454 pKa = 10.58RR455 pKa = 11.84QQEE458 pKa = 4.26LIKK461 pKa = 10.26WVSDD465 pKa = 3.46SMNKK469 pKa = 9.75HH470 pKa = 6.08PMGQIYY476 pKa = 9.68RR477 pKa = 11.84VVRR480 pKa = 11.84EE481 pKa = 4.12ALDD484 pKa = 4.0AEE486 pKa = 4.68SDD488 pKa = 3.86SSSSSDD494 pKa = 2.92SDD496 pKa = 3.39
MM1 pKa = 7.73SDD3 pKa = 3.66YY4 pKa = 11.3NPDD7 pKa = 3.21VLQQLVKK14 pKa = 10.94DD15 pKa = 4.5GIKK18 pKa = 9.87TIEE21 pKa = 4.64LLQQSPEE28 pKa = 4.27DD29 pKa = 3.66FQKK32 pKa = 10.02TYY34 pKa = 10.56GRR36 pKa = 11.84SAIQEE41 pKa = 3.9PSTRR45 pKa = 11.84ARR47 pKa = 11.84IQSWEE52 pKa = 4.05SRR54 pKa = 11.84NPDD57 pKa = 2.98HH58 pKa = 8.22DD59 pKa = 3.87YY60 pKa = 11.2TGNKK64 pKa = 7.4NQRR67 pKa = 11.84SEE69 pKa = 4.25GAKK72 pKa = 9.54EE73 pKa = 3.59RR74 pKa = 11.84ANKK77 pKa = 10.16SEE79 pKa = 4.19SAGTASADD87 pKa = 3.54GGHH90 pKa = 6.72GDD92 pKa = 3.68KK93 pKa = 10.73PSNNGGDD100 pKa = 3.65SQNEE104 pKa = 4.14YY105 pKa = 10.4QGSDD109 pKa = 3.36QQVWDD114 pKa = 3.49AAYY117 pKa = 11.07NDD119 pKa = 4.47GNSGGAWGGPTGGLPTAGEE138 pKa = 3.97RR139 pKa = 11.84GYY141 pKa = 9.98PITTGNQEE149 pKa = 4.06FEE151 pKa = 4.82GYY153 pKa = 10.34HH154 pKa = 6.45PDD156 pKa = 3.57GPVDD160 pKa = 3.27ARR162 pKa = 11.84EE163 pKa = 3.96YY164 pKa = 10.21NQISSMDD171 pKa = 3.72HH172 pKa = 5.74EE173 pKa = 4.57MSAAEE178 pKa = 4.06TLGSSTQLTTMRR190 pKa = 11.84NATTDD195 pKa = 3.38DD196 pKa = 3.37FAKK199 pKa = 10.19IFEE202 pKa = 4.48EE203 pKa = 4.86GTPKK207 pKa = 9.59VHH209 pKa = 6.94RR210 pKa = 11.84RR211 pKa = 11.84LRR213 pKa = 11.84GITAVVPAQKK223 pKa = 10.33QPGAVGGPVKK233 pKa = 10.57KK234 pKa = 9.83GTDD237 pKa = 3.22GSTASTLLGDD247 pKa = 3.97VPLSGSGAIPNVHH260 pKa = 7.08PSLLHH265 pKa = 4.98QPKK268 pKa = 10.34KK269 pKa = 10.18NAHH272 pKa = 6.42AEE274 pKa = 4.0NAQGSVQDD282 pKa = 3.7VSTTGAIGQSDD293 pKa = 3.88EE294 pKa = 4.43AAHH297 pKa = 6.78FDD299 pKa = 4.02QEE301 pKa = 4.88IEE303 pKa = 4.23GKK305 pKa = 10.62LNLVLKK311 pKa = 10.3EE312 pKa = 4.38LDD314 pKa = 4.63LISKK318 pKa = 10.15KK319 pKa = 10.38LDD321 pKa = 3.35YY322 pKa = 10.97LPEE325 pKa = 5.38IKK327 pKa = 10.58EE328 pKa = 4.09EE329 pKa = 3.65IRR331 pKa = 11.84NINKK335 pKa = 10.34KK336 pKa = 8.04ITNLSLGLSTVEE348 pKa = 4.21NYY350 pKa = 9.38IKK352 pKa = 11.05SMMIIIPSSGKK363 pKa = 9.59PDD365 pKa = 3.5VNDD368 pKa = 3.66TAEE371 pKa = 4.08VNPDD375 pKa = 3.29LKK377 pKa = 11.16AVIGRR382 pKa = 11.84DD383 pKa = 3.0KK384 pKa = 11.08TRR386 pKa = 11.84GLKK389 pKa = 9.92EE390 pKa = 4.15VTTHH394 pKa = 7.59RR395 pKa = 11.84SDD397 pKa = 5.14LEE399 pKa = 4.16SLDD402 pKa = 4.21LLSPATSEE410 pKa = 4.3IDD412 pKa = 3.66PKK414 pKa = 11.45YY415 pKa = 9.84LTQDD419 pKa = 3.09LDD421 pKa = 3.99FTKK424 pKa = 10.93SNAANFVPGNDD435 pKa = 2.96ASSFYY440 pKa = 10.54TIVAMIKK447 pKa = 10.89GEE449 pKa = 4.07VTDD452 pKa = 3.54VKK454 pKa = 10.58RR455 pKa = 11.84QQEE458 pKa = 4.26LIKK461 pKa = 10.26WVSDD465 pKa = 3.46SMNKK469 pKa = 9.75HH470 pKa = 6.08PMGQIYY476 pKa = 9.68RR477 pKa = 11.84VVRR480 pKa = 11.84EE481 pKa = 4.12ALDD484 pKa = 4.0AEE486 pKa = 4.68SDD488 pKa = 3.86SSSSSDD494 pKa = 2.92SDD496 pKa = 3.39
Molecular weight: 53.6 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q287X8|Q287X8_9MONO Isoform of Q287Y0 V protein OS=Beilong virus OX=341053 GN=P/V/C PE=4 SV=1
MM1 pKa = 7.5SSSSLSRR8 pKa = 11.84MEE10 pKa = 4.03SRR12 pKa = 11.84LSSFYY17 pKa = 11.07SRR19 pKa = 11.84VRR21 pKa = 11.84KK22 pKa = 7.18TFRR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84TAEE30 pKa = 3.76VPSRR34 pKa = 11.84SRR36 pKa = 11.84RR37 pKa = 11.84QEE39 pKa = 3.83PEE41 pKa = 4.03SKK43 pKa = 10.26VGSPAIPIMITPVIKK58 pKa = 10.11IRR60 pKa = 11.84EE61 pKa = 4.03ARR63 pKa = 11.84EE64 pKa = 3.76LKK66 pKa = 10.45RR67 pKa = 11.84EE68 pKa = 3.96QTRR71 pKa = 11.84ARR73 pKa = 11.84ARR75 pKa = 11.84EE76 pKa = 3.95LLQLMEE82 pKa = 5.12DD83 pKa = 3.91MEE85 pKa = 5.28INPQITGVTPKK96 pKa = 10.06TSTKK100 pKa = 10.47GQINRR105 pKa = 11.84YY106 pKa = 8.12GMLRR110 pKa = 11.84IMMGIVAEE118 pKa = 4.31LGEE121 pKa = 4.49VPQVDD126 pKa = 4.82FPRR129 pKa = 11.84LEE131 pKa = 4.01KK132 pKa = 10.83EE133 pKa = 4.67GILSQQEE140 pKa = 4.06IKK142 pKa = 10.71NLRR145 pKa = 11.84DD146 pKa = 3.7TIPTVQLMLEE156 pKa = 4.35SIIRR160 pKa = 3.68
MM1 pKa = 7.5SSSSLSRR8 pKa = 11.84MEE10 pKa = 4.03SRR12 pKa = 11.84LSSFYY17 pKa = 11.07SRR19 pKa = 11.84VRR21 pKa = 11.84KK22 pKa = 7.18TFRR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84TAEE30 pKa = 3.76VPSRR34 pKa = 11.84SRR36 pKa = 11.84RR37 pKa = 11.84QEE39 pKa = 3.83PEE41 pKa = 4.03SKK43 pKa = 10.26VGSPAIPIMITPVIKK58 pKa = 10.11IRR60 pKa = 11.84EE61 pKa = 4.03ARR63 pKa = 11.84EE64 pKa = 3.76LKK66 pKa = 10.45RR67 pKa = 11.84EE68 pKa = 3.96QTRR71 pKa = 11.84ARR73 pKa = 11.84ARR75 pKa = 11.84EE76 pKa = 3.95LLQLMEE82 pKa = 5.12DD83 pKa = 3.91MEE85 pKa = 5.28INPQITGVTPKK96 pKa = 10.06TSTKK100 pKa = 10.47GQINRR105 pKa = 11.84YY106 pKa = 8.12GMLRR110 pKa = 11.84IMMGIVAEE118 pKa = 4.31LGEE121 pKa = 4.49VPQVDD126 pKa = 4.82FPRR129 pKa = 11.84LEE131 pKa = 4.01KK132 pKa = 10.83EE133 pKa = 4.67GILSQQEE140 pKa = 4.06IKK142 pKa = 10.71NLRR145 pKa = 11.84DD146 pKa = 3.7TIPTVQLMLEE156 pKa = 4.35SIIRR160 pKa = 3.68
Molecular weight: 18.44 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
5903 |
76 |
2172 |
536.6 |
60.14 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.166 ± 0.69 | 1.745 ± 0.322 |
5.319 ± 0.448 | 5.658 ± 0.408 |
3.371 ± 0.441 | 6.437 ± 0.85 |
1.999 ± 0.303 | 6.844 ± 0.485 |
5.607 ± 0.368 | 9.08 ± 0.941 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.965 ± 0.223 | 5.438 ± 0.348 |
4.794 ± 0.501 | 3.761 ± 0.383 |
5.336 ± 0.505 | 7.86 ± 0.485 |
6.37 ± 0.418 | 6.166 ± 0.385 |
1.067 ± 0.103 | 4.015 ± 0.39 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
