
Haloarcula sp. JP-L23
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Halobacteriales; Haloarculaceae; Haloarcula; unclassified Haloarcula
Average proteome isoelectric point is 4.87
Get precalculated fractions of proteins
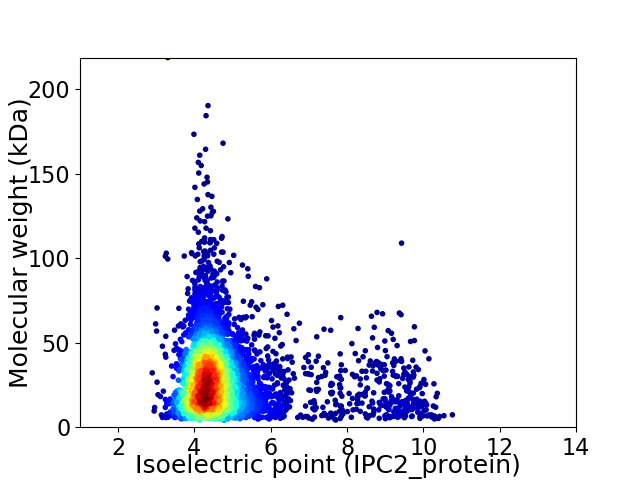
Virtual 2D-PAGE plot for 4757 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6G8TAZ4|A0A6G8TAZ4_9EURY Uncharacterized protein OS=Haloarcula sp. JP-L23 OX=2716717 GN=G9465_17025 PE=4 SV=1
MM1 pKa = 7.66ISGTGVRR8 pKa = 11.84SLLLQTGIGPIDD20 pKa = 4.5AIIGLFVSIVLGIVNFAAQVLPTVLLALLVLGVGYY55 pKa = 10.38VVADD59 pKa = 4.09RR60 pKa = 11.84LDD62 pKa = 3.5TFVFEE67 pKa = 4.45WALEE71 pKa = 3.85NDD73 pKa = 3.79VDD75 pKa = 4.6GKK77 pKa = 11.06AGDD80 pKa = 4.17TPASTVVADD89 pKa = 3.98EE90 pKa = 5.27DD91 pKa = 4.53GVATAAGQLVRR102 pKa = 11.84YY103 pKa = 8.64LVLVVAVVAAIRR115 pKa = 11.84VLNLSEE121 pKa = 4.23LQEE124 pKa = 4.36LLDD127 pKa = 3.66IVIGYY132 pKa = 8.42VPNVVAAAVLLVVGFGVGRR151 pKa = 11.84IVRR154 pKa = 11.84SLVPGLVDD162 pKa = 3.45RR163 pKa = 11.84TDD165 pKa = 4.09LGDD168 pKa = 3.94SFPTTHH174 pKa = 7.4FGRR177 pKa = 11.84VVDD180 pKa = 4.21ADD182 pKa = 3.58GGTVGRR188 pKa = 11.84LAGIAVEE195 pKa = 4.28YY196 pKa = 10.34YY197 pKa = 9.95VYY199 pKa = 10.96LVTLYY204 pKa = 10.69VVAATLAIGSVAGVLGAAVSYY225 pKa = 11.21VPTLLGALVLLLLGAIVADD244 pKa = 3.78YY245 pKa = 10.82VGTVVRR251 pKa = 11.84GVDD254 pKa = 3.88EE255 pKa = 4.52VADD258 pKa = 4.02SPFEE262 pKa = 4.06SLVTGGVQAVVYY274 pKa = 9.74LFTVVIALDD283 pKa = 3.67VAGVDD288 pKa = 3.53ATLLGLLLLAVVLPVGLAFALAFGLSVGYY317 pKa = 10.36GGQDD321 pKa = 3.07YY322 pKa = 10.96VEE324 pKa = 4.6QYY326 pKa = 11.17LGTDD330 pKa = 3.47EE331 pKa = 4.86AAEE334 pKa = 4.03
MM1 pKa = 7.66ISGTGVRR8 pKa = 11.84SLLLQTGIGPIDD20 pKa = 4.5AIIGLFVSIVLGIVNFAAQVLPTVLLALLVLGVGYY55 pKa = 10.38VVADD59 pKa = 4.09RR60 pKa = 11.84LDD62 pKa = 3.5TFVFEE67 pKa = 4.45WALEE71 pKa = 3.85NDD73 pKa = 3.79VDD75 pKa = 4.6GKK77 pKa = 11.06AGDD80 pKa = 4.17TPASTVVADD89 pKa = 3.98EE90 pKa = 5.27DD91 pKa = 4.53GVATAAGQLVRR102 pKa = 11.84YY103 pKa = 8.64LVLVVAVVAAIRR115 pKa = 11.84VLNLSEE121 pKa = 4.23LQEE124 pKa = 4.36LLDD127 pKa = 3.66IVIGYY132 pKa = 8.42VPNVVAAAVLLVVGFGVGRR151 pKa = 11.84IVRR154 pKa = 11.84SLVPGLVDD162 pKa = 3.45RR163 pKa = 11.84TDD165 pKa = 4.09LGDD168 pKa = 3.94SFPTTHH174 pKa = 7.4FGRR177 pKa = 11.84VVDD180 pKa = 4.21ADD182 pKa = 3.58GGTVGRR188 pKa = 11.84LAGIAVEE195 pKa = 4.28YY196 pKa = 10.34YY197 pKa = 9.95VYY199 pKa = 10.96LVTLYY204 pKa = 10.69VVAATLAIGSVAGVLGAAVSYY225 pKa = 11.21VPTLLGALVLLLLGAIVADD244 pKa = 3.78YY245 pKa = 10.82VGTVVRR251 pKa = 11.84GVDD254 pKa = 3.88EE255 pKa = 4.52VADD258 pKa = 4.02SPFEE262 pKa = 4.06SLVTGGVQAVVYY274 pKa = 9.74LFTVVIALDD283 pKa = 3.67VAGVDD288 pKa = 3.53ATLLGLLLLAVVLPVGLAFALAFGLSVGYY317 pKa = 10.36GGQDD321 pKa = 3.07YY322 pKa = 10.96VEE324 pKa = 4.6QYY326 pKa = 11.17LGTDD330 pKa = 3.47EE331 pKa = 4.86AAEE334 pKa = 4.03
Molecular weight: 34.15 kDa
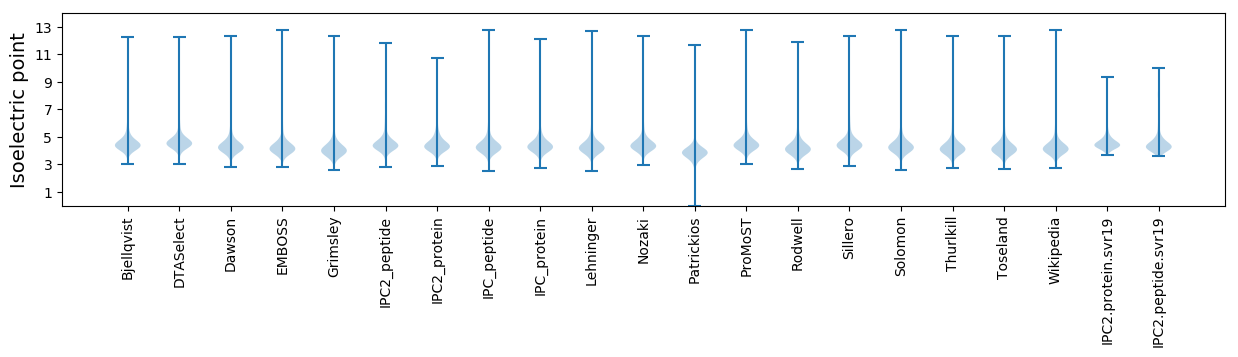
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6G8TF09|A0A6G8TF09_9EURY Uncharacterized protein OS=Haloarcula sp. JP-L23 OX=2716717 GN=G9465_24110 PE=4 SV=1
MM1 pKa = 7.26SRR3 pKa = 11.84KK4 pKa = 9.88LYY6 pKa = 10.15FVEE9 pKa = 4.23LAEE12 pKa = 4.26TEE14 pKa = 4.24RR15 pKa = 11.84AEE17 pKa = 4.9LEE19 pKa = 4.21ALVSAGNHH27 pKa = 5.26SAQKK31 pKa = 8.92LTRR34 pKa = 11.84ARR36 pKa = 11.84ILLKK40 pKa = 10.33SDD42 pKa = 4.26EE43 pKa = 4.41GWSDD47 pKa = 3.4PKK49 pKa = 10.73IADD52 pKa = 3.8ALDD55 pKa = 3.73CGRR58 pKa = 11.84ATVQRR63 pKa = 11.84TRR65 pKa = 11.84EE66 pKa = 3.79RR67 pKa = 11.84FSRR70 pKa = 11.84QRR72 pKa = 11.84LGAIEE77 pKa = 4.31RR78 pKa = 11.84KK79 pKa = 9.81NPAVSPRR86 pKa = 11.84ANSTATPRR94 pKa = 11.84PIDD97 pKa = 3.47CVGVQRR103 pKa = 11.84AARR106 pKa = 11.84RR107 pKa = 11.84ARR109 pKa = 11.84SLDD112 pKa = 3.1ASPPRR117 pKa = 11.84RR118 pKa = 11.84HH119 pKa = 6.28ACHH122 pKa = 6.16PQQ124 pKa = 2.76
MM1 pKa = 7.26SRR3 pKa = 11.84KK4 pKa = 9.88LYY6 pKa = 10.15FVEE9 pKa = 4.23LAEE12 pKa = 4.26TEE14 pKa = 4.24RR15 pKa = 11.84AEE17 pKa = 4.9LEE19 pKa = 4.21ALVSAGNHH27 pKa = 5.26SAQKK31 pKa = 8.92LTRR34 pKa = 11.84ARR36 pKa = 11.84ILLKK40 pKa = 10.33SDD42 pKa = 4.26EE43 pKa = 4.41GWSDD47 pKa = 3.4PKK49 pKa = 10.73IADD52 pKa = 3.8ALDD55 pKa = 3.73CGRR58 pKa = 11.84ATVQRR63 pKa = 11.84TRR65 pKa = 11.84EE66 pKa = 3.79RR67 pKa = 11.84FSRR70 pKa = 11.84QRR72 pKa = 11.84LGAIEE77 pKa = 4.31RR78 pKa = 11.84KK79 pKa = 9.81NPAVSPRR86 pKa = 11.84ANSTATPRR94 pKa = 11.84PIDD97 pKa = 3.47CVGVQRR103 pKa = 11.84AARR106 pKa = 11.84RR107 pKa = 11.84ARR109 pKa = 11.84SLDD112 pKa = 3.1ASPPRR117 pKa = 11.84RR118 pKa = 11.84HH119 pKa = 6.28ACHH122 pKa = 6.16PQQ124 pKa = 2.76
Molecular weight: 13.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1338033 |
36 |
2113 |
281.3 |
30.59 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.633 ± 0.045 | 0.75 ± 0.011 |
8.459 ± 0.053 | 8.105 ± 0.056 |
3.298 ± 0.024 | 8.343 ± 0.041 |
2.051 ± 0.018 | 3.874 ± 0.026 |
1.868 ± 0.022 | 8.878 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.821 ± 0.015 | 2.42 ± 0.023 |
4.663 ± 0.025 | 2.795 ± 0.022 |
6.331 ± 0.035 | 5.622 ± 0.032 |
6.85 ± 0.041 | 9.279 ± 0.042 |
1.186 ± 0.016 | 2.774 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
