
Shahe hepe-like virus 2
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.26
Get precalculated fractions of proteins
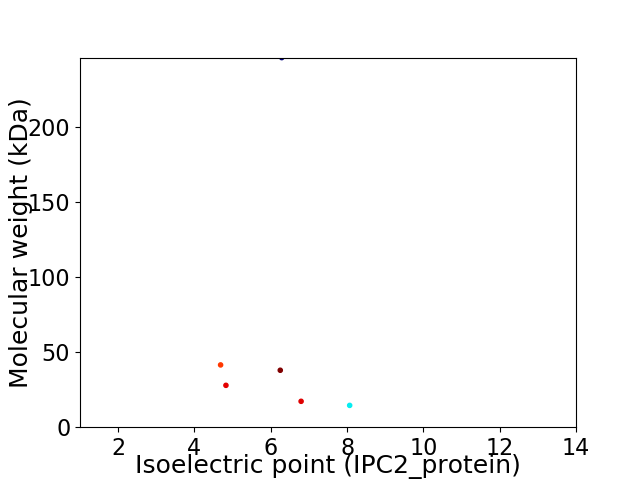
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KK59|A0A1L3KK59_9VIRU Alphavirus-like MT domain-containing protein OS=Shahe hepe-like virus 2 OX=1923416 PE=4 SV=1
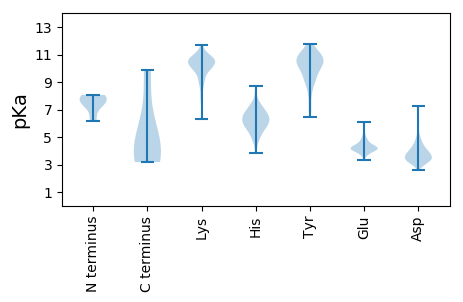
MM1 pKa = 7.33NFDD4 pKa = 5.02LIEE7 pKa = 4.44DD8 pKa = 3.97VVGDD12 pKa = 3.43VMNIYY17 pKa = 10.1PEE19 pKa = 4.14QPIHH23 pKa = 6.07ARR25 pKa = 11.84AMIKK29 pKa = 10.05RR30 pKa = 11.84HH31 pKa = 4.77FTLEE35 pKa = 3.83KK36 pKa = 10.75LGDD39 pKa = 3.84HH40 pKa = 7.0AAGKK44 pKa = 9.85YY45 pKa = 7.18WLIPITFQEE54 pKa = 4.41PDD56 pKa = 2.87QHH58 pKa = 8.32AEE60 pKa = 3.57YY61 pKa = 10.48MMYY64 pKa = 10.44RR65 pKa = 11.84ISNYY69 pKa = 9.75KK70 pKa = 10.57LEE72 pKa = 4.41FNNTKK77 pKa = 10.05HH78 pKa = 5.46QMNVSTTASNNNDD91 pKa = 3.02GYY93 pKa = 11.36FRR95 pKa = 11.84IAWIRR100 pKa = 11.84DD101 pKa = 3.47PMIGTPKK108 pKa = 10.64DD109 pKa = 3.31YY110 pKa = 10.77MNFVTRR116 pKa = 11.84LPGSKK121 pKa = 9.92LVSWYY126 pKa = 10.46DD127 pKa = 3.35NLSFQVEE134 pKa = 4.6TNGWRR139 pKa = 11.84YY140 pKa = 9.26CKK142 pKa = 10.03KK143 pKa = 10.8GKK145 pKa = 9.96DD146 pKa = 3.86KK147 pKa = 11.17RR148 pKa = 11.84LTDD151 pKa = 3.57YY152 pKa = 11.65GDD154 pKa = 3.66IIIAKK159 pKa = 9.99FSYY162 pKa = 9.89TGLDD166 pKa = 3.95GVWTEE171 pKa = 4.47SFDD174 pKa = 3.64IEE176 pKa = 4.64YY177 pKa = 10.48SVPTLPSKK185 pKa = 7.86WTPITCYY192 pKa = 10.96ALDD195 pKa = 3.63TLAAAGAADD204 pKa = 4.12YY205 pKa = 11.29KK206 pKa = 11.28DD207 pKa = 4.31LLSFTASYY215 pKa = 10.99SGAQQLYY222 pKa = 10.0CNFKK226 pKa = 10.79LGLEE230 pKa = 4.16KK231 pKa = 10.77GAIPTQKK238 pKa = 10.1KK239 pKa = 10.29GRR241 pKa = 11.84ITLGKK246 pKa = 8.8PVVIALIFTATPVSGDD262 pKa = 3.78PYY264 pKa = 10.69DD265 pKa = 3.56VTYY268 pKa = 10.31EE269 pKa = 4.16FPFTSAEE276 pKa = 4.18CEE278 pKa = 4.34SIGTGLWGGNLDD290 pKa = 4.81ASLLDD295 pKa = 2.99IEE297 pKa = 5.92FYY299 pKa = 10.67INPYY303 pKa = 10.07GEE305 pKa = 4.67LGADD309 pKa = 3.55VLSLKK314 pKa = 10.33SVYY317 pKa = 9.33WDD319 pKa = 3.17QDD321 pKa = 3.6PNISADD327 pKa = 3.11EE328 pKa = 4.09RR329 pKa = 11.84MSLKK333 pKa = 10.6AQLIFAEE340 pKa = 4.67EE341 pKa = 4.42VPDD344 pKa = 5.01SINDD348 pKa = 4.17LIEE351 pKa = 4.04TLNIGAGSAYY361 pKa = 9.74EE362 pKa = 4.18DD363 pKa = 4.53LVNKK367 pKa = 9.4YY368 pKa = 9.85
MM1 pKa = 7.33NFDD4 pKa = 5.02LIEE7 pKa = 4.44DD8 pKa = 3.97VVGDD12 pKa = 3.43VMNIYY17 pKa = 10.1PEE19 pKa = 4.14QPIHH23 pKa = 6.07ARR25 pKa = 11.84AMIKK29 pKa = 10.05RR30 pKa = 11.84HH31 pKa = 4.77FTLEE35 pKa = 3.83KK36 pKa = 10.75LGDD39 pKa = 3.84HH40 pKa = 7.0AAGKK44 pKa = 9.85YY45 pKa = 7.18WLIPITFQEE54 pKa = 4.41PDD56 pKa = 2.87QHH58 pKa = 8.32AEE60 pKa = 3.57YY61 pKa = 10.48MMYY64 pKa = 10.44RR65 pKa = 11.84ISNYY69 pKa = 9.75KK70 pKa = 10.57LEE72 pKa = 4.41FNNTKK77 pKa = 10.05HH78 pKa = 5.46QMNVSTTASNNNDD91 pKa = 3.02GYY93 pKa = 11.36FRR95 pKa = 11.84IAWIRR100 pKa = 11.84DD101 pKa = 3.47PMIGTPKK108 pKa = 10.64DD109 pKa = 3.31YY110 pKa = 10.77MNFVTRR116 pKa = 11.84LPGSKK121 pKa = 9.92LVSWYY126 pKa = 10.46DD127 pKa = 3.35NLSFQVEE134 pKa = 4.6TNGWRR139 pKa = 11.84YY140 pKa = 9.26CKK142 pKa = 10.03KK143 pKa = 10.8GKK145 pKa = 9.96DD146 pKa = 3.86KK147 pKa = 11.17RR148 pKa = 11.84LTDD151 pKa = 3.57YY152 pKa = 11.65GDD154 pKa = 3.66IIIAKK159 pKa = 9.99FSYY162 pKa = 9.89TGLDD166 pKa = 3.95GVWTEE171 pKa = 4.47SFDD174 pKa = 3.64IEE176 pKa = 4.64YY177 pKa = 10.48SVPTLPSKK185 pKa = 7.86WTPITCYY192 pKa = 10.96ALDD195 pKa = 3.63TLAAAGAADD204 pKa = 4.12YY205 pKa = 11.29KK206 pKa = 11.28DD207 pKa = 4.31LLSFTASYY215 pKa = 10.99SGAQQLYY222 pKa = 10.0CNFKK226 pKa = 10.79LGLEE230 pKa = 4.16KK231 pKa = 10.77GAIPTQKK238 pKa = 10.1KK239 pKa = 10.29GRR241 pKa = 11.84ITLGKK246 pKa = 8.8PVVIALIFTATPVSGDD262 pKa = 3.78PYY264 pKa = 10.69DD265 pKa = 3.56VTYY268 pKa = 10.31EE269 pKa = 4.16FPFTSAEE276 pKa = 4.18CEE278 pKa = 4.34SIGTGLWGGNLDD290 pKa = 4.81ASLLDD295 pKa = 2.99IEE297 pKa = 5.92FYY299 pKa = 10.67INPYY303 pKa = 10.07GEE305 pKa = 4.67LGADD309 pKa = 3.55VLSLKK314 pKa = 10.33SVYY317 pKa = 9.33WDD319 pKa = 3.17QDD321 pKa = 3.6PNISADD327 pKa = 3.11EE328 pKa = 4.09RR329 pKa = 11.84MSLKK333 pKa = 10.6AQLIFAEE340 pKa = 4.67EE341 pKa = 4.42VPDD344 pKa = 5.01SINDD348 pKa = 4.17LIEE351 pKa = 4.04TLNIGAGSAYY361 pKa = 9.74EE362 pKa = 4.18DD363 pKa = 4.53LVNKK367 pKa = 9.4YY368 pKa = 9.85
Molecular weight: 41.47 kDa
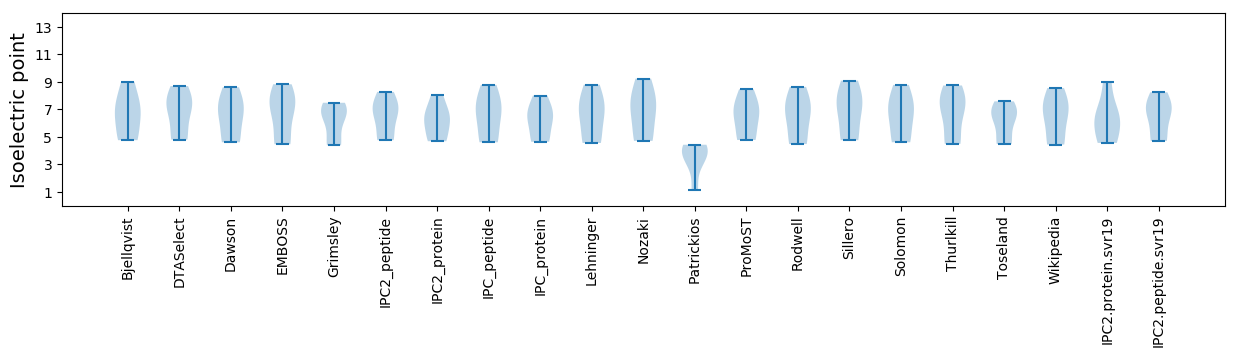
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KK79|A0A1L3KK79_9VIRU Uncharacterized protein OS=Shahe hepe-like virus 2 OX=1923416 PE=4 SV=1
MM1 pKa = 7.55SKK3 pKa = 10.84GFTLDD8 pKa = 2.63SYY10 pKa = 11.7YY11 pKa = 11.26LFLPLSFLGAKK22 pKa = 9.76FCLVLIFQSRR32 pKa = 11.84FVSVISKK39 pKa = 9.4FISLSLFYY47 pKa = 11.34VNFSSCQSSTTGFSLICYY65 pKa = 9.87SEE67 pKa = 5.51AILDD71 pKa = 4.02LSWIISISTLRR82 pKa = 11.84LPTEE86 pKa = 4.0CWMVQHH92 pKa = 7.24APHH95 pKa = 5.96QQPKK99 pKa = 8.51CTSTNFGMTSTQTGYY114 pKa = 10.15PKK116 pKa = 11.03LLTCHH121 pKa = 6.22GRR123 pKa = 11.84WTMKK127 pKa = 10.58LL128 pKa = 3.32
MM1 pKa = 7.55SKK3 pKa = 10.84GFTLDD8 pKa = 2.63SYY10 pKa = 11.7YY11 pKa = 11.26LFLPLSFLGAKK22 pKa = 9.76FCLVLIFQSRR32 pKa = 11.84FVSVISKK39 pKa = 9.4FISLSLFYY47 pKa = 11.34VNFSSCQSSTTGFSLICYY65 pKa = 9.87SEE67 pKa = 5.51AILDD71 pKa = 4.02LSWIISISTLRR82 pKa = 11.84LPTEE86 pKa = 4.0CWMVQHH92 pKa = 7.24APHH95 pKa = 5.96QQPKK99 pKa = 8.51CTSTNFGMTSTQTGYY114 pKa = 10.15PKK116 pKa = 11.03LLTCHH121 pKa = 6.22GRR123 pKa = 11.84WTMKK127 pKa = 10.58LL128 pKa = 3.32
Molecular weight: 14.52 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3403 |
128 |
2173 |
567.2 |
64.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.464 ± 0.414 | 1.822 ± 0.355 |
5.877 ± 0.734 | 6.142 ± 0.533 |
5.73 ± 0.806 | 5.348 ± 0.357 |
2.557 ± 0.497 | 7.141 ± 0.257 |
7.229 ± 0.63 | 7.787 ± 0.656 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.056 ± 0.209 | 4.731 ± 0.282 |
4.731 ± 0.609 | 3.438 ± 0.403 |
3.879 ± 0.515 | 5.789 ± 1.093 |
6.759 ± 0.357 | 5.671 ± 0.29 |
1.058 ± 0.281 | 3.791 ± 0.55 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
