
Capybara microvirus Cap1_SP_161
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.08
Get precalculated fractions of proteins
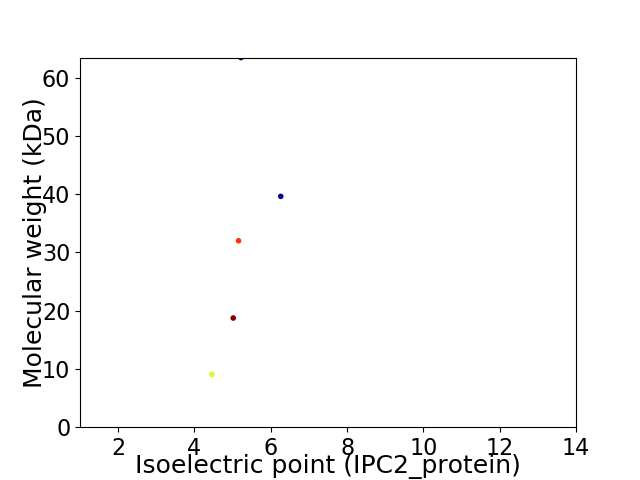
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W4G3|A0A4P8W4G3_9VIRU Replication initiator protein OS=Capybara microvirus Cap1_SP_161 OX=2585397 PE=4 SV=1
MM1 pKa = 7.31NLYY4 pKa = 10.34VLYY7 pKa = 10.45DD8 pKa = 3.69KK9 pKa = 10.37EE10 pKa = 4.1AHH12 pKa = 6.41IVQCPYY18 pKa = 10.24PQPLEE23 pKa = 3.84NDD25 pKa = 4.03TIARR29 pKa = 11.84RR30 pKa = 11.84WWKK33 pKa = 10.46KK34 pKa = 6.75STEE37 pKa = 4.13SLVDD41 pKa = 3.17VMGILPSMISLYY53 pKa = 10.91CVGIWNKK60 pKa = 7.95EE61 pKa = 4.02TGEE64 pKa = 3.87ITPCFDD70 pKa = 3.09EE71 pKa = 4.49VAVISSDD78 pKa = 3.73DD79 pKa = 3.28
MM1 pKa = 7.31NLYY4 pKa = 10.34VLYY7 pKa = 10.45DD8 pKa = 3.69KK9 pKa = 10.37EE10 pKa = 4.1AHH12 pKa = 6.41IVQCPYY18 pKa = 10.24PQPLEE23 pKa = 3.84NDD25 pKa = 4.03TIARR29 pKa = 11.84RR30 pKa = 11.84WWKK33 pKa = 10.46KK34 pKa = 6.75STEE37 pKa = 4.13SLVDD41 pKa = 3.17VMGILPSMISLYY53 pKa = 10.91CVGIWNKK60 pKa = 7.95EE61 pKa = 4.02TGEE64 pKa = 3.87ITPCFDD70 pKa = 3.09EE71 pKa = 4.49VAVISSDD78 pKa = 3.73DD79 pKa = 3.28
Molecular weight: 9.07 kDa
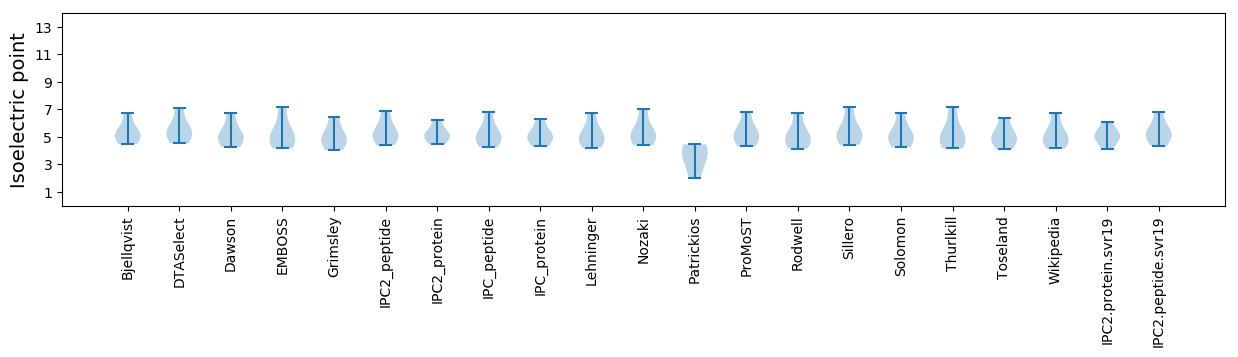
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W5A4|A0A4P8W5A4_9VIRU Major capsid protein OS=Capybara microvirus Cap1_SP_161 OX=2585397 PE=3 SV=1
MM1 pKa = 7.36ICSSPKK7 pKa = 10.27LVYY10 pKa = 10.59LKK12 pKa = 10.07NNCSIRR18 pKa = 11.84VPCGHH23 pKa = 7.46CINCRR28 pKa = 11.84IQKK31 pKa = 10.05SSDD34 pKa = 2.37WATRR38 pKa = 11.84ICLEE42 pKa = 4.61LPFWDD47 pKa = 3.87SNIFTTLTYY56 pKa = 10.98DD57 pKa = 4.2DD58 pKa = 4.92DD59 pKa = 5.97HH60 pKa = 8.43IPSDD64 pKa = 3.59WSIHH68 pKa = 6.67KK69 pKa = 10.47DD70 pKa = 3.26DD71 pKa = 4.14LQKK74 pKa = 10.67FFKK77 pKa = 10.37RR78 pKa = 11.84LRR80 pKa = 11.84RR81 pKa = 11.84EE82 pKa = 3.26IEE84 pKa = 3.99YY85 pKa = 10.11NAKK88 pKa = 10.61SNGLEE93 pKa = 4.12VPKK96 pKa = 10.26IAYY99 pKa = 7.62FASGEE104 pKa = 4.14YY105 pKa = 10.02GDD107 pKa = 4.4KK108 pKa = 10.87SMRR111 pKa = 11.84PHH113 pKa = 4.89YY114 pKa = 9.89HH115 pKa = 6.84AIIFGVGLDD124 pKa = 3.56LNNYY128 pKa = 9.04NNEE131 pKa = 4.01LQCYY135 pKa = 9.83YY136 pKa = 10.7SDD138 pKa = 4.68CKK140 pKa = 10.87LIEE143 pKa = 4.46NNWSYY148 pKa = 11.71GNVYY152 pKa = 9.76NGCVSYY158 pKa = 11.29DD159 pKa = 3.49SARR162 pKa = 11.84YY163 pKa = 6.4TADD166 pKa = 3.33YY167 pKa = 11.03VLKK170 pKa = 10.46QYY172 pKa = 10.94SGEE175 pKa = 4.11LKK177 pKa = 10.49QKK179 pKa = 10.5VYY181 pKa = 10.46TDD183 pKa = 3.75NGLEE187 pKa = 4.2TPFCIHH193 pKa = 6.82SNGIGKK199 pKa = 9.68RR200 pKa = 11.84FVEE203 pKa = 5.05VYY205 pKa = 10.05EE206 pKa = 4.1SDD208 pKa = 3.77IIGKK212 pKa = 9.89GYY214 pKa = 10.22IIIRR218 pKa = 11.84GVKK221 pKa = 9.38KK222 pKa = 10.15ALPRR226 pKa = 11.84YY227 pKa = 9.64FYY229 pKa = 10.73DD230 pKa = 3.58ILNKK234 pKa = 9.81KK235 pKa = 8.21YY236 pKa = 9.62RR237 pKa = 11.84WHH239 pKa = 6.01KK240 pKa = 10.77CEE242 pKa = 4.08YY243 pKa = 10.71DD244 pKa = 3.87DD245 pKa = 5.23DD246 pKa = 5.77GKK248 pKa = 10.92MIFGFSYY255 pKa = 10.6FEE257 pKa = 3.97NFQKK261 pKa = 10.65HH262 pKa = 3.91MKK264 pKa = 9.37NYY266 pKa = 9.31KK267 pKa = 10.24GEE269 pKa = 4.06YY270 pKa = 10.05LEE272 pKa = 4.53ADD274 pKa = 3.1QMEE277 pKa = 5.06LFRR280 pKa = 11.84RR281 pKa = 11.84WRR283 pKa = 11.84DD284 pKa = 3.3EE285 pKa = 3.92CKK287 pKa = 10.29EE288 pKa = 4.52LYY290 pKa = 10.63DD291 pKa = 4.86CDD293 pKa = 4.2VSFDD297 pKa = 4.9DD298 pKa = 5.49FLQKK302 pKa = 10.38EE303 pKa = 4.25KK304 pKa = 11.09CDD306 pKa = 4.13FSDD309 pKa = 5.11KK310 pKa = 10.67IEE312 pKa = 4.17QNVARR317 pKa = 11.84NKK319 pKa = 10.46LNEE322 pKa = 3.94HH323 pKa = 6.54KK324 pKa = 10.63KK325 pKa = 10.62IIDD328 pKa = 3.6AQRR331 pKa = 11.84SLII334 pKa = 3.94
MM1 pKa = 7.36ICSSPKK7 pKa = 10.27LVYY10 pKa = 10.59LKK12 pKa = 10.07NNCSIRR18 pKa = 11.84VPCGHH23 pKa = 7.46CINCRR28 pKa = 11.84IQKK31 pKa = 10.05SSDD34 pKa = 2.37WATRR38 pKa = 11.84ICLEE42 pKa = 4.61LPFWDD47 pKa = 3.87SNIFTTLTYY56 pKa = 10.98DD57 pKa = 4.2DD58 pKa = 4.92DD59 pKa = 5.97HH60 pKa = 8.43IPSDD64 pKa = 3.59WSIHH68 pKa = 6.67KK69 pKa = 10.47DD70 pKa = 3.26DD71 pKa = 4.14LQKK74 pKa = 10.67FFKK77 pKa = 10.37RR78 pKa = 11.84LRR80 pKa = 11.84RR81 pKa = 11.84EE82 pKa = 3.26IEE84 pKa = 3.99YY85 pKa = 10.11NAKK88 pKa = 10.61SNGLEE93 pKa = 4.12VPKK96 pKa = 10.26IAYY99 pKa = 7.62FASGEE104 pKa = 4.14YY105 pKa = 10.02GDD107 pKa = 4.4KK108 pKa = 10.87SMRR111 pKa = 11.84PHH113 pKa = 4.89YY114 pKa = 9.89HH115 pKa = 6.84AIIFGVGLDD124 pKa = 3.56LNNYY128 pKa = 9.04NNEE131 pKa = 4.01LQCYY135 pKa = 9.83YY136 pKa = 10.7SDD138 pKa = 4.68CKK140 pKa = 10.87LIEE143 pKa = 4.46NNWSYY148 pKa = 11.71GNVYY152 pKa = 9.76NGCVSYY158 pKa = 11.29DD159 pKa = 3.49SARR162 pKa = 11.84YY163 pKa = 6.4TADD166 pKa = 3.33YY167 pKa = 11.03VLKK170 pKa = 10.46QYY172 pKa = 10.94SGEE175 pKa = 4.11LKK177 pKa = 10.49QKK179 pKa = 10.5VYY181 pKa = 10.46TDD183 pKa = 3.75NGLEE187 pKa = 4.2TPFCIHH193 pKa = 6.82SNGIGKK199 pKa = 9.68RR200 pKa = 11.84FVEE203 pKa = 5.05VYY205 pKa = 10.05EE206 pKa = 4.1SDD208 pKa = 3.77IIGKK212 pKa = 9.89GYY214 pKa = 10.22IIIRR218 pKa = 11.84GVKK221 pKa = 9.38KK222 pKa = 10.15ALPRR226 pKa = 11.84YY227 pKa = 9.64FYY229 pKa = 10.73DD230 pKa = 3.58ILNKK234 pKa = 9.81KK235 pKa = 8.21YY236 pKa = 9.62RR237 pKa = 11.84WHH239 pKa = 6.01KK240 pKa = 10.77CEE242 pKa = 4.08YY243 pKa = 10.71DD244 pKa = 3.87DD245 pKa = 5.23DD246 pKa = 5.77GKK248 pKa = 10.92MIFGFSYY255 pKa = 10.6FEE257 pKa = 3.97NFQKK261 pKa = 10.65HH262 pKa = 3.91MKK264 pKa = 9.37NYY266 pKa = 9.31KK267 pKa = 10.24GEE269 pKa = 4.06YY270 pKa = 10.05LEE272 pKa = 4.53ADD274 pKa = 3.1QMEE277 pKa = 5.06LFRR280 pKa = 11.84RR281 pKa = 11.84WRR283 pKa = 11.84DD284 pKa = 3.3EE285 pKa = 3.92CKK287 pKa = 10.29EE288 pKa = 4.52LYY290 pKa = 10.63DD291 pKa = 4.86CDD293 pKa = 4.2VSFDD297 pKa = 4.9DD298 pKa = 5.49FLQKK302 pKa = 10.38EE303 pKa = 4.25KK304 pKa = 11.09CDD306 pKa = 4.13FSDD309 pKa = 5.11KK310 pKa = 10.67IEE312 pKa = 4.17QNVARR317 pKa = 11.84NKK319 pKa = 10.46LNEE322 pKa = 3.94HH323 pKa = 6.54KK324 pKa = 10.63KK325 pKa = 10.62IIDD328 pKa = 3.6AQRR331 pKa = 11.84SLII334 pKa = 3.94
Molecular weight: 39.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1425 |
79 |
558 |
285.0 |
32.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.053 ± 0.72 | 1.825 ± 0.627 |
7.509 ± 0.825 | 5.614 ± 0.556 |
4.912 ± 0.695 | 5.895 ± 0.531 |
1.684 ± 0.398 | 5.263 ± 0.73 |
6.877 ± 1.527 | 8.07 ± 0.531 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.456 ± 0.278 | 5.895 ± 0.498 |
3.368 ± 0.955 | 5.053 ± 1.018 |
4.07 ± 0.409 | 9.404 ± 0.718 |
4.14 ± 0.822 | 5.965 ± 0.632 |
1.544 ± 0.307 | 5.404 ± 0.831 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
