
Sanxia atyid shrimp virus 1
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.09
Get precalculated fractions of proteins
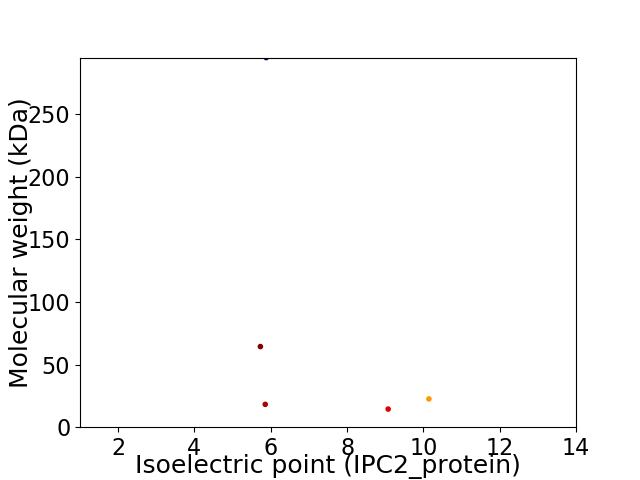
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KK18|A0A1L3KK18_9VIRU Uncharacterized protein OS=Sanxia atyid shrimp virus 1 OX=1923355 PE=4 SV=1
MM1 pKa = 7.43NFLTASKK8 pKa = 7.36TTLALIWLRR17 pKa = 11.84IAPMAIFWIVSSVVRR32 pKa = 11.84LSVTTVCPNQLLNRR46 pKa = 11.84LNTLNSLFPVAISVVLHH63 pKa = 5.14VLRR66 pKa = 11.84FFLTLLSHH74 pKa = 5.98VNILILMLVIRR85 pKa = 11.84GLEE88 pKa = 3.85FSSLIFSLITSCPLSLLLYY107 pKa = 10.14YY108 pKa = 11.1LLLFLLAVSPAFLLAAAVANKK129 pKa = 10.01LFSPSSATTCVFMALLPLVTSDD151 pKa = 4.86ALFLSSSSVHH161 pKa = 6.42FNDD164 pKa = 2.98ITQGSISPSPSVPSGQWFLTSPLTMLNPEE193 pKa = 4.25CSYY196 pKa = 11.09ILSTSRR202 pKa = 11.84IWPSEE207 pKa = 3.92CEE209 pKa = 4.2LPLSCGDD216 pKa = 3.61MLEE219 pKa = 4.2KK220 pKa = 10.56QSLGGAFFSEE230 pKa = 4.06TDD232 pKa = 2.99LCFRR236 pKa = 11.84FFHH239 pKa = 7.54DD240 pKa = 3.22YY241 pKa = 9.2HH242 pKa = 6.51TFQSYY247 pKa = 8.83IVYY250 pKa = 10.23GILEE254 pKa = 4.36GTLNTSSYY262 pKa = 11.1QIDD265 pKa = 3.94FHH267 pKa = 9.37SFDD270 pKa = 3.8PPIWLHH276 pKa = 5.9SPSARR281 pKa = 11.84RR282 pKa = 11.84SGWIHH287 pKa = 7.07LDD289 pKa = 3.01AAHH292 pKa = 6.56YY293 pKa = 8.93FLHH296 pKa = 6.86YY297 pKa = 10.78DD298 pKa = 3.52PLKK301 pKa = 11.08NKK303 pKa = 9.04YY304 pKa = 9.6AWDD307 pKa = 3.51SSLFNTKK314 pKa = 10.09RR315 pKa = 11.84EE316 pKa = 3.98CHH318 pKa = 5.47MVNTSAVFFEE328 pKa = 5.07APYY331 pKa = 10.35IDD333 pKa = 4.76YY334 pKa = 7.73NTSSLLLHH342 pKa = 6.9DD343 pKa = 6.43LIFQDD348 pKa = 4.51YY349 pKa = 9.44LSSGIPLNYY358 pKa = 9.03TLHH361 pKa = 6.44PKK363 pKa = 8.25KK364 pKa = 9.05TYY366 pKa = 10.96AFGTEE371 pKa = 4.07LVLSFVPNSYY381 pKa = 10.6FSFSVLGNGFISSDD395 pKa = 3.75SFTHH399 pKa = 6.77LLDD402 pKa = 3.8NGTYY406 pKa = 8.35ITIEE410 pKa = 4.14SDD412 pKa = 3.57PEE414 pKa = 4.02TLSISLYY421 pKa = 10.22PLDD424 pKa = 4.32FNPFVEE430 pKa = 3.82IFEE433 pKa = 4.51YY434 pKa = 10.31KK435 pKa = 9.63GHH437 pKa = 6.34SFSFMCTSTCAVEE450 pKa = 4.18FLHH453 pKa = 5.67YY454 pKa = 10.03TNILSWLNLTSTSIKK469 pKa = 8.51NTKK472 pKa = 7.72EE473 pKa = 3.46RR474 pKa = 11.84CVRR477 pKa = 11.84SISKK481 pKa = 10.07VSYY484 pKa = 8.54TNFVTGIVYY493 pKa = 10.38RR494 pKa = 11.84SISTFEE500 pKa = 3.84HH501 pKa = 5.49AVIRR505 pKa = 11.84LFRR508 pKa = 11.84LFVDD512 pKa = 5.47AISTAFQQLLTYY524 pKa = 8.93VLSMEE529 pKa = 4.21RR530 pKa = 11.84QFKK533 pKa = 10.32IIEE536 pKa = 4.09FGIILALCTAHH547 pKa = 6.64FNNISTALLLTLSIWYY563 pKa = 7.65VAPFNTQQ570 pKa = 2.34
MM1 pKa = 7.43NFLTASKK8 pKa = 7.36TTLALIWLRR17 pKa = 11.84IAPMAIFWIVSSVVRR32 pKa = 11.84LSVTTVCPNQLLNRR46 pKa = 11.84LNTLNSLFPVAISVVLHH63 pKa = 5.14VLRR66 pKa = 11.84FFLTLLSHH74 pKa = 5.98VNILILMLVIRR85 pKa = 11.84GLEE88 pKa = 3.85FSSLIFSLITSCPLSLLLYY107 pKa = 10.14YY108 pKa = 11.1LLLFLLAVSPAFLLAAAVANKK129 pKa = 10.01LFSPSSATTCVFMALLPLVTSDD151 pKa = 4.86ALFLSSSSVHH161 pKa = 6.42FNDD164 pKa = 2.98ITQGSISPSPSVPSGQWFLTSPLTMLNPEE193 pKa = 4.25CSYY196 pKa = 11.09ILSTSRR202 pKa = 11.84IWPSEE207 pKa = 3.92CEE209 pKa = 4.2LPLSCGDD216 pKa = 3.61MLEE219 pKa = 4.2KK220 pKa = 10.56QSLGGAFFSEE230 pKa = 4.06TDD232 pKa = 2.99LCFRR236 pKa = 11.84FFHH239 pKa = 7.54DD240 pKa = 3.22YY241 pKa = 9.2HH242 pKa = 6.51TFQSYY247 pKa = 8.83IVYY250 pKa = 10.23GILEE254 pKa = 4.36GTLNTSSYY262 pKa = 11.1QIDD265 pKa = 3.94FHH267 pKa = 9.37SFDD270 pKa = 3.8PPIWLHH276 pKa = 5.9SPSARR281 pKa = 11.84RR282 pKa = 11.84SGWIHH287 pKa = 7.07LDD289 pKa = 3.01AAHH292 pKa = 6.56YY293 pKa = 8.93FLHH296 pKa = 6.86YY297 pKa = 10.78DD298 pKa = 3.52PLKK301 pKa = 11.08NKK303 pKa = 9.04YY304 pKa = 9.6AWDD307 pKa = 3.51SSLFNTKK314 pKa = 10.09RR315 pKa = 11.84EE316 pKa = 3.98CHH318 pKa = 5.47MVNTSAVFFEE328 pKa = 5.07APYY331 pKa = 10.35IDD333 pKa = 4.76YY334 pKa = 7.73NTSSLLLHH342 pKa = 6.9DD343 pKa = 6.43LIFQDD348 pKa = 4.51YY349 pKa = 9.44LSSGIPLNYY358 pKa = 9.03TLHH361 pKa = 6.44PKK363 pKa = 8.25KK364 pKa = 9.05TYY366 pKa = 10.96AFGTEE371 pKa = 4.07LVLSFVPNSYY381 pKa = 10.6FSFSVLGNGFISSDD395 pKa = 3.75SFTHH399 pKa = 6.77LLDD402 pKa = 3.8NGTYY406 pKa = 8.35ITIEE410 pKa = 4.14SDD412 pKa = 3.57PEE414 pKa = 4.02TLSISLYY421 pKa = 10.22PLDD424 pKa = 4.32FNPFVEE430 pKa = 3.82IFEE433 pKa = 4.51YY434 pKa = 10.31KK435 pKa = 9.63GHH437 pKa = 6.34SFSFMCTSTCAVEE450 pKa = 4.18FLHH453 pKa = 5.67YY454 pKa = 10.03TNILSWLNLTSTSIKK469 pKa = 8.51NTKK472 pKa = 7.72EE473 pKa = 3.46RR474 pKa = 11.84CVRR477 pKa = 11.84SISKK481 pKa = 10.07VSYY484 pKa = 8.54TNFVTGIVYY493 pKa = 10.38RR494 pKa = 11.84SISTFEE500 pKa = 3.84HH501 pKa = 5.49AVIRR505 pKa = 11.84LFRR508 pKa = 11.84LFVDD512 pKa = 5.47AISTAFQQLLTYY524 pKa = 8.93VLSMEE529 pKa = 4.21RR530 pKa = 11.84QFKK533 pKa = 10.32IIEE536 pKa = 4.09FGIILALCTAHH547 pKa = 6.64FNNISTALLLTLSIWYY563 pKa = 7.65VAPFNTQQ570 pKa = 2.34
Molecular weight: 64.36 kDa
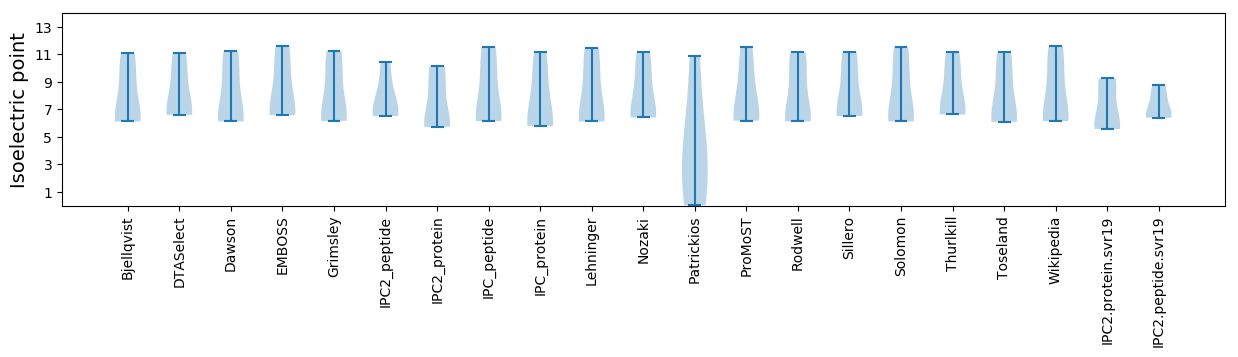
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KKA1|A0A1L3KKA1_9VIRU Helicase OS=Sanxia atyid shrimp virus 1 OX=1923355 PE=4 SV=1
MM1 pKa = 7.95ADD3 pKa = 3.05TAKK6 pKa = 10.47RR7 pKa = 11.84PRR9 pKa = 11.84FFTRR13 pKa = 11.84PPPMMPRR20 pKa = 11.84GPPPQARR27 pKa = 11.84KK28 pKa = 9.09SARR31 pKa = 11.84SRR33 pKa = 11.84KK34 pKa = 9.22NGQSSDD40 pKa = 3.57FFEE43 pKa = 4.91GVLDD47 pKa = 3.81ALRR50 pKa = 11.84RR51 pKa = 11.84TSSQPDD57 pKa = 3.88VLCLLALALLVAYY70 pKa = 9.56DD71 pKa = 3.31ITHH74 pKa = 7.24IDD76 pKa = 2.94RR77 pKa = 11.84LAAFFSARR85 pKa = 11.84TSTTQLGSWISANSQVIAGLVILLPGLYY113 pKa = 9.85KK114 pKa = 10.68FPVKK118 pKa = 10.56SRR120 pKa = 11.84FIAGIVTFVTVLALPSQSSYY140 pKa = 11.34VYY142 pKa = 9.62FAVSLGMTVFWQVRR156 pKa = 11.84QPGIRR161 pKa = 11.84LVLFSALLLYY171 pKa = 10.67LYY173 pKa = 9.84VFNGAALGISPTTSATPPPVTTTTASTTTRR203 pKa = 11.84PPAGRR208 pKa = 3.68
MM1 pKa = 7.95ADD3 pKa = 3.05TAKK6 pKa = 10.47RR7 pKa = 11.84PRR9 pKa = 11.84FFTRR13 pKa = 11.84PPPMMPRR20 pKa = 11.84GPPPQARR27 pKa = 11.84KK28 pKa = 9.09SARR31 pKa = 11.84SRR33 pKa = 11.84KK34 pKa = 9.22NGQSSDD40 pKa = 3.57FFEE43 pKa = 4.91GVLDD47 pKa = 3.81ALRR50 pKa = 11.84RR51 pKa = 11.84TSSQPDD57 pKa = 3.88VLCLLALALLVAYY70 pKa = 9.56DD71 pKa = 3.31ITHH74 pKa = 7.24IDD76 pKa = 2.94RR77 pKa = 11.84LAAFFSARR85 pKa = 11.84TSTTQLGSWISANSQVIAGLVILLPGLYY113 pKa = 9.85KK114 pKa = 10.68FPVKK118 pKa = 10.56SRR120 pKa = 11.84FIAGIVTFVTVLALPSQSSYY140 pKa = 11.34VYY142 pKa = 9.62FAVSLGMTVFWQVRR156 pKa = 11.84QPGIRR161 pKa = 11.84LVLFSALLLYY171 pKa = 10.67LYY173 pKa = 9.84VFNGAALGISPTTSATPPPVTTTTASTTTRR203 pKa = 11.84PPAGRR208 pKa = 3.68
Molecular weight: 22.55 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3699 |
128 |
2629 |
739.8 |
82.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.921 ± 0.686 | 2.379 ± 0.685 |
6.164 ± 1.489 | 3.839 ± 0.629 |
5.758 ± 0.89 | 4.38 ± 0.445 |
2.703 ± 0.335 | 6.11 ± 0.443 |
4.866 ± 1.221 | 10.192 ± 1.614 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.974 ± 0.265 | 4.028 ± 0.435 |
5.569 ± 0.565 | 2.595 ± 0.494 |
4.677 ± 0.702 | 9.651 ± 1.041 |
5.434 ± 1.214 | 7.651 ± 0.728 |
1.027 ± 0.237 | 4.082 ± 0.279 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
