
Epstein-Barr virus (strain B95-8) (HHV-4) (Human herpesvirus 4)
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Peploviricota; Herviviricetes; Herpesvirales; Herpesviridae; Gammaherpesvirinae; Lymphocryptovirus; Human gammaherpesvirus 4
Average proteome isoelectric point is 7.14
Get precalculated fractions of proteins
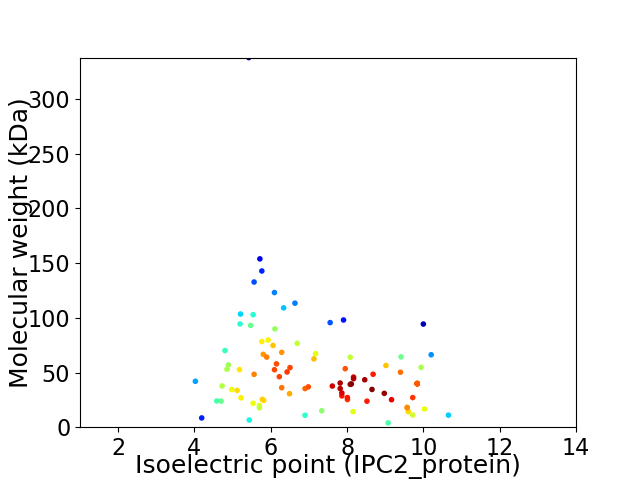
Virtual 2D-PAGE plot for 95 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P03231|GH_EBVB9 Envelope glycoprotein H OS=Epstein-Barr virus (strain B95-8) OX=10377 GN=gH PE=1 SV=1
MM1 pKa = 7.25EE2 pKa = 6.06HH3 pKa = 7.2DD4 pKa = 4.84LEE6 pKa = 5.12RR7 pKa = 11.84GPPGPRR13 pKa = 11.84RR14 pKa = 11.84PPRR17 pKa = 11.84GPPLSSSLGLALLLLLLALLFWLYY41 pKa = 10.78IVMSDD46 pKa = 3.13WTGGALLVLYY56 pKa = 10.32SFALMLIIIILIIFIFRR73 pKa = 11.84RR74 pKa = 11.84DD75 pKa = 3.73LLCPLGALCILLLMITLLLIALWNLHH101 pKa = 4.99GQALFLGIVLFIFGCLLVLGIWIYY125 pKa = 10.72LLEE128 pKa = 4.15MLWRR132 pKa = 11.84LGATIWQLLAFFLAFFLDD150 pKa = 5.18LILLIIALYY159 pKa = 9.75LQQNWWTLLVDD170 pKa = 5.12LLWLLLFLAILIWMYY185 pKa = 10.53YY186 pKa = 8.99HH187 pKa = 6.85GQRR190 pKa = 11.84HH191 pKa = 5.39SDD193 pKa = 3.35EE194 pKa = 4.43HH195 pKa = 7.75HH196 pKa = 6.96HH197 pKa = 7.46DD198 pKa = 4.28DD199 pKa = 5.28SLPHH203 pKa = 6.25PQQATDD209 pKa = 3.84DD210 pKa = 4.24SGHH213 pKa = 6.22EE214 pKa = 4.08SDD216 pKa = 4.7SNSNEE221 pKa = 3.73GRR223 pKa = 11.84HH224 pKa = 5.73HH225 pKa = 6.63LLVSGAGDD233 pKa = 4.11GPPLCSQNLGAPGGGPDD250 pKa = 5.41NGPQDD255 pKa = 4.7PDD257 pKa = 3.47NTDD260 pKa = 3.56DD261 pKa = 5.97NGPQDD266 pKa = 5.0PDD268 pKa = 3.47NTDD271 pKa = 3.47DD272 pKa = 5.12NGPHH276 pKa = 7.25DD277 pKa = 5.7PLPQDD282 pKa = 4.74PDD284 pKa = 3.54NTDD287 pKa = 3.56DD288 pKa = 5.97NGPQDD293 pKa = 5.0PDD295 pKa = 3.47NTDD298 pKa = 3.47DD299 pKa = 5.12NGPHH303 pKa = 7.22DD304 pKa = 5.4PLPHH308 pKa = 6.43SPSDD312 pKa = 3.48SAGNDD317 pKa = 3.34GGPPQLTEE325 pKa = 3.78EE326 pKa = 4.43VEE328 pKa = 4.41NKK330 pKa = 10.57GGDD333 pKa = 3.37QGPPLMTDD341 pKa = 3.13GGGGHH346 pKa = 5.9SHH348 pKa = 7.88DD349 pKa = 4.68SGHH352 pKa = 6.96GGGDD356 pKa = 3.16PHH358 pKa = 8.56LPTLLLGSSGSGGDD372 pKa = 4.23DD373 pKa = 4.27DD374 pKa = 6.1DD375 pKa = 4.11PHH377 pKa = 8.7GPVQLSYY384 pKa = 11.75YY385 pKa = 10.41DD386 pKa = 3.38
MM1 pKa = 7.25EE2 pKa = 6.06HH3 pKa = 7.2DD4 pKa = 4.84LEE6 pKa = 5.12RR7 pKa = 11.84GPPGPRR13 pKa = 11.84RR14 pKa = 11.84PPRR17 pKa = 11.84GPPLSSSLGLALLLLLLALLFWLYY41 pKa = 10.78IVMSDD46 pKa = 3.13WTGGALLVLYY56 pKa = 10.32SFALMLIIIILIIFIFRR73 pKa = 11.84RR74 pKa = 11.84DD75 pKa = 3.73LLCPLGALCILLLMITLLLIALWNLHH101 pKa = 4.99GQALFLGIVLFIFGCLLVLGIWIYY125 pKa = 10.72LLEE128 pKa = 4.15MLWRR132 pKa = 11.84LGATIWQLLAFFLAFFLDD150 pKa = 5.18LILLIIALYY159 pKa = 9.75LQQNWWTLLVDD170 pKa = 5.12LLWLLLFLAILIWMYY185 pKa = 10.53YY186 pKa = 8.99HH187 pKa = 6.85GQRR190 pKa = 11.84HH191 pKa = 5.39SDD193 pKa = 3.35EE194 pKa = 4.43HH195 pKa = 7.75HH196 pKa = 6.96HH197 pKa = 7.46DD198 pKa = 4.28DD199 pKa = 5.28SLPHH203 pKa = 6.25PQQATDD209 pKa = 3.84DD210 pKa = 4.24SGHH213 pKa = 6.22EE214 pKa = 4.08SDD216 pKa = 4.7SNSNEE221 pKa = 3.73GRR223 pKa = 11.84HH224 pKa = 5.73HH225 pKa = 6.63LLVSGAGDD233 pKa = 4.11GPPLCSQNLGAPGGGPDD250 pKa = 5.41NGPQDD255 pKa = 4.7PDD257 pKa = 3.47NTDD260 pKa = 3.56DD261 pKa = 5.97NGPQDD266 pKa = 5.0PDD268 pKa = 3.47NTDD271 pKa = 3.47DD272 pKa = 5.12NGPHH276 pKa = 7.25DD277 pKa = 5.7PLPQDD282 pKa = 4.74PDD284 pKa = 3.54NTDD287 pKa = 3.56DD288 pKa = 5.97NGPQDD293 pKa = 5.0PDD295 pKa = 3.47NTDD298 pKa = 3.47DD299 pKa = 5.12NGPHH303 pKa = 7.22DD304 pKa = 5.4PLPHH308 pKa = 6.43SPSDD312 pKa = 3.48SAGNDD317 pKa = 3.34GGPPQLTEE325 pKa = 3.78EE326 pKa = 4.43VEE328 pKa = 4.41NKK330 pKa = 10.57GGDD333 pKa = 3.37QGPPLMTDD341 pKa = 3.13GGGGHH346 pKa = 5.9SHH348 pKa = 7.88DD349 pKa = 4.68SGHH352 pKa = 6.96GGGDD356 pKa = 3.16PHH358 pKa = 8.56LPTLLLGSSGSGGDD372 pKa = 4.23DD373 pKa = 4.27DD374 pKa = 6.1DD375 pKa = 4.11PHH377 pKa = 8.7GPVQLSYY384 pKa = 11.75YY385 pKa = 10.41DD386 pKa = 3.38
Molecular weight: 41.98 kDa
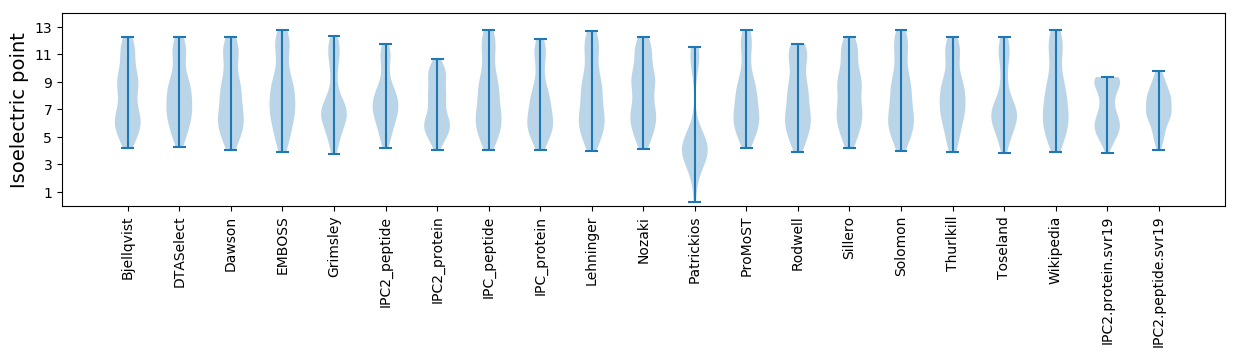
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P25939|TRM1_EBVB9 Tripartite terminase subunit 1 OS=Epstein-Barr virus (strain B95-8) OX=10377 GN=TRM1 PE=3 SV=2
MM1 pKa = 7.61LAHH4 pKa = 7.23LNQVTRR10 pKa = 11.84IPPCPPFSGRR20 pKa = 11.84EE21 pKa = 3.88ARR23 pKa = 11.84LKK25 pKa = 10.34FHH27 pKa = 6.85FFSWSTFMLSWPNNATLRR45 pKa = 11.84EE46 pKa = 3.95IRR48 pKa = 11.84TRR50 pKa = 11.84AATNLTHH57 pKa = 6.93HH58 pKa = 6.79PHH60 pKa = 6.72LVDD63 pKa = 3.51TLYY66 pKa = 10.67HH67 pKa = 7.07ASPQTPFLTRR77 pKa = 11.84SGALYY82 pKa = 10.56RR83 pKa = 11.84FVTCCNCTLPNISIQQCKK101 pKa = 10.23AGDD104 pKa = 3.57RR105 pKa = 11.84PGDD108 pKa = 3.69LEE110 pKa = 6.7IILQSNGGGRR120 pKa = 11.84PASFQFPSSPTGSLLRR136 pKa = 11.84CIVAASLLPEE146 pKa = 4.09VSVGHH151 pKa = 5.82QEE153 pKa = 4.19LSPLRR158 pKa = 11.84SRR160 pKa = 11.84SQGGQTDD167 pKa = 3.87VRR169 pKa = 11.84SGPDD173 pKa = 3.1PARR176 pKa = 11.84RR177 pKa = 11.84LVALLRR183 pKa = 11.84RR184 pKa = 11.84EE185 pKa = 4.23DD186 pKa = 4.14GAPKK190 pKa = 10.26DD191 pKa = 3.82PPLGPFGHH199 pKa = 7.02PRR201 pKa = 11.84GPGPAKK207 pKa = 10.74SEE209 pKa = 4.16DD210 pKa = 3.81EE211 pKa = 3.97EE212 pKa = 4.85SEE214 pKa = 4.53RR215 pKa = 11.84RR216 pKa = 11.84DD217 pKa = 3.74APPPPLDD224 pKa = 3.71SSFQASRR231 pKa = 11.84LVPVGPGFRR240 pKa = 11.84LLVFNTNRR248 pKa = 11.84VINTKK253 pKa = 9.58LVCSEE258 pKa = 4.65PLVKK262 pKa = 9.9MRR264 pKa = 11.84VCNVPRR270 pKa = 11.84LINNFVARR278 pKa = 11.84KK279 pKa = 8.91YY280 pKa = 9.69VVKK283 pKa = 9.23EE284 pKa = 3.69TAFTVSLFFTDD295 pKa = 4.55GVGANLAINVNISGTYY311 pKa = 10.4LSFLLAMTSLRR322 pKa = 11.84CFLPVEE328 pKa = 5.02AIYY331 pKa = 9.19PAAVSNWNSTLDD343 pKa = 3.49LHH345 pKa = 6.85GLEE348 pKa = 4.26NQSLVRR354 pKa = 11.84EE355 pKa = 4.13NRR357 pKa = 11.84SGVFWTTNFPSVVSCRR373 pKa = 11.84DD374 pKa = 3.27GLNVSWFKK382 pKa = 11.05AATATISRR390 pKa = 11.84VHH392 pKa = 5.72GQTLEE397 pKa = 3.64QHH399 pKa = 7.74LIRR402 pKa = 11.84EE403 pKa = 4.04ITPIVTHH410 pKa = 6.79RR411 pKa = 11.84EE412 pKa = 3.93AKK414 pKa = 9.45ISRR417 pKa = 11.84IKK419 pKa = 10.86NRR421 pKa = 11.84LFTLLEE427 pKa = 3.94LRR429 pKa = 11.84NRR431 pKa = 11.84SQIQVLHH438 pKa = 6.05KK439 pKa = 10.55RR440 pKa = 11.84FLEE443 pKa = 4.21GLLDD447 pKa = 4.03CASLLRR453 pKa = 11.84LDD455 pKa = 4.26PSCINRR461 pKa = 11.84IASEE465 pKa = 4.21GLFDD469 pKa = 4.39FSKK472 pKa = 10.9RR473 pKa = 11.84SIAHH477 pKa = 5.1SKK479 pKa = 9.41NRR481 pKa = 11.84HH482 pKa = 4.08EE483 pKa = 4.59CALLGHH489 pKa = 6.33RR490 pKa = 11.84HH491 pKa = 5.44SANVTKK497 pKa = 10.75LVVNEE502 pKa = 4.0RR503 pKa = 11.84KK504 pKa = 7.87TRR506 pKa = 11.84LDD508 pKa = 3.04ILGRR512 pKa = 11.84NANFLTRR519 pKa = 11.84CKK521 pKa = 10.32HH522 pKa = 4.71QVNLRR527 pKa = 11.84QSPIFLTLLRR537 pKa = 11.84HH538 pKa = 4.86IRR540 pKa = 11.84RR541 pKa = 11.84RR542 pKa = 11.84LGLGRR547 pKa = 11.84ASVKK551 pKa = 10.64RR552 pKa = 11.84EE553 pKa = 3.54ITLLLAHH560 pKa = 6.77LRR562 pKa = 11.84KK563 pKa = 8.68KK564 pKa = 7.99TAPIHH569 pKa = 6.52CRR571 pKa = 11.84DD572 pKa = 3.63AQVV575 pKa = 2.73
MM1 pKa = 7.61LAHH4 pKa = 7.23LNQVTRR10 pKa = 11.84IPPCPPFSGRR20 pKa = 11.84EE21 pKa = 3.88ARR23 pKa = 11.84LKK25 pKa = 10.34FHH27 pKa = 6.85FFSWSTFMLSWPNNATLRR45 pKa = 11.84EE46 pKa = 3.95IRR48 pKa = 11.84TRR50 pKa = 11.84AATNLTHH57 pKa = 6.93HH58 pKa = 6.79PHH60 pKa = 6.72LVDD63 pKa = 3.51TLYY66 pKa = 10.67HH67 pKa = 7.07ASPQTPFLTRR77 pKa = 11.84SGALYY82 pKa = 10.56RR83 pKa = 11.84FVTCCNCTLPNISIQQCKK101 pKa = 10.23AGDD104 pKa = 3.57RR105 pKa = 11.84PGDD108 pKa = 3.69LEE110 pKa = 6.7IILQSNGGGRR120 pKa = 11.84PASFQFPSSPTGSLLRR136 pKa = 11.84CIVAASLLPEE146 pKa = 4.09VSVGHH151 pKa = 5.82QEE153 pKa = 4.19LSPLRR158 pKa = 11.84SRR160 pKa = 11.84SQGGQTDD167 pKa = 3.87VRR169 pKa = 11.84SGPDD173 pKa = 3.1PARR176 pKa = 11.84RR177 pKa = 11.84LVALLRR183 pKa = 11.84RR184 pKa = 11.84EE185 pKa = 4.23DD186 pKa = 4.14GAPKK190 pKa = 10.26DD191 pKa = 3.82PPLGPFGHH199 pKa = 7.02PRR201 pKa = 11.84GPGPAKK207 pKa = 10.74SEE209 pKa = 4.16DD210 pKa = 3.81EE211 pKa = 3.97EE212 pKa = 4.85SEE214 pKa = 4.53RR215 pKa = 11.84RR216 pKa = 11.84DD217 pKa = 3.74APPPPLDD224 pKa = 3.71SSFQASRR231 pKa = 11.84LVPVGPGFRR240 pKa = 11.84LLVFNTNRR248 pKa = 11.84VINTKK253 pKa = 9.58LVCSEE258 pKa = 4.65PLVKK262 pKa = 9.9MRR264 pKa = 11.84VCNVPRR270 pKa = 11.84LINNFVARR278 pKa = 11.84KK279 pKa = 8.91YY280 pKa = 9.69VVKK283 pKa = 9.23EE284 pKa = 3.69TAFTVSLFFTDD295 pKa = 4.55GVGANLAINVNISGTYY311 pKa = 10.4LSFLLAMTSLRR322 pKa = 11.84CFLPVEE328 pKa = 5.02AIYY331 pKa = 9.19PAAVSNWNSTLDD343 pKa = 3.49LHH345 pKa = 6.85GLEE348 pKa = 4.26NQSLVRR354 pKa = 11.84EE355 pKa = 4.13NRR357 pKa = 11.84SGVFWTTNFPSVVSCRR373 pKa = 11.84DD374 pKa = 3.27GLNVSWFKK382 pKa = 11.05AATATISRR390 pKa = 11.84VHH392 pKa = 5.72GQTLEE397 pKa = 3.64QHH399 pKa = 7.74LIRR402 pKa = 11.84EE403 pKa = 4.04ITPIVTHH410 pKa = 6.79RR411 pKa = 11.84EE412 pKa = 3.93AKK414 pKa = 9.45ISRR417 pKa = 11.84IKK419 pKa = 10.86NRR421 pKa = 11.84LFTLLEE427 pKa = 3.94LRR429 pKa = 11.84NRR431 pKa = 11.84SQIQVLHH438 pKa = 6.05KK439 pKa = 10.55RR440 pKa = 11.84FLEE443 pKa = 4.21GLLDD447 pKa = 4.03CASLLRR453 pKa = 11.84LDD455 pKa = 4.26PSCINRR461 pKa = 11.84IASEE465 pKa = 4.21GLFDD469 pKa = 4.39FSKK472 pKa = 10.9RR473 pKa = 11.84SIAHH477 pKa = 5.1SKK479 pKa = 9.41NRR481 pKa = 11.84HH482 pKa = 4.08EE483 pKa = 4.59CALLGHH489 pKa = 6.33RR490 pKa = 11.84HH491 pKa = 5.44SANVTKK497 pKa = 10.75LVVNEE502 pKa = 4.0RR503 pKa = 11.84KK504 pKa = 7.87TRR506 pKa = 11.84LDD508 pKa = 3.04ILGRR512 pKa = 11.84NANFLTRR519 pKa = 11.84CKK521 pKa = 10.32HH522 pKa = 4.71QVNLRR527 pKa = 11.84QSPIFLTLLRR537 pKa = 11.84HH538 pKa = 4.86IRR540 pKa = 11.84RR541 pKa = 11.84RR542 pKa = 11.84LGLGRR547 pKa = 11.84ASVKK551 pKa = 10.64RR552 pKa = 11.84EE553 pKa = 3.54ITLLLAHH560 pKa = 6.77LRR562 pKa = 11.84KK563 pKa = 8.68KK564 pKa = 7.99TAPIHH569 pKa = 6.52CRR571 pKa = 11.84DD572 pKa = 3.63AQVV575 pKa = 2.73
Molecular weight: 64.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
46367 |
39 |
3149 |
488.1 |
53.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.356 ± 0.48 | 1.945 ± 0.141 |
4.331 ± 0.162 | 5.137 ± 0.18 |
3.384 ± 0.208 | 7.906 ± 0.563 |
2.519 ± 0.095 | 3.311 ± 0.198 |
2.892 ± 0.169 | 10.05 ± 0.387 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.799 ± 0.122 | 2.741 ± 0.2 |
9.66 ± 0.766 | 4.096 ± 0.167 |
7.1 ± 0.34 | 7.697 ± 0.202 |
6.004 ± 0.316 | 6.33 ± 0.248 |
1.184 ± 0.065 | 2.558 ± 0.157 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
