
Alteromonas phage vB_AcoS-R7M
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; Queuovirinae; Amoyvirus; Alteromonas virus R7M
Average proteome isoelectric point is 6.02
Get precalculated fractions of proteins
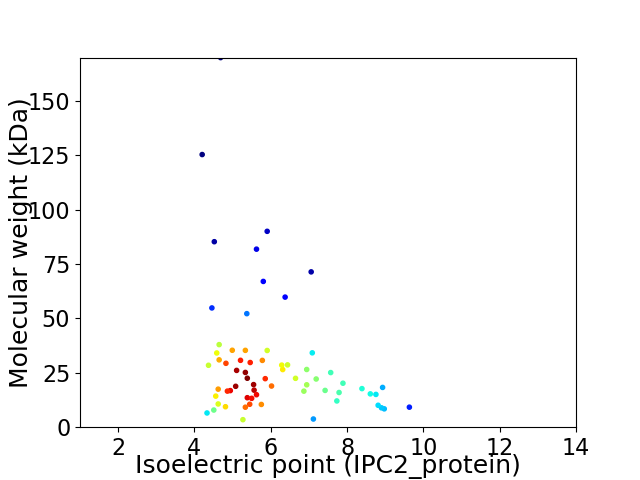
Virtual 2D-PAGE plot for 67 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6M3YNF1|A0A6M3YNF1_9CAUD DNA helicase/restriction enzyme OS=Alteromonas phage vB_AcoS-R7M OX=2729541 GN=vBAcoSR7M_53 PE=4 SV=1
MM1 pKa = 7.72AVTTLDD7 pKa = 3.54LVQTNGTPLPSEE19 pKa = 4.05LTVLEE24 pKa = 4.74GEE26 pKa = 4.37VEE28 pKa = 4.4YY29 pKa = 11.33NGSGLVAATAPAALNFNHH47 pKa = 7.52KK48 pKa = 9.09DD49 pKa = 3.4TAIVSYY55 pKa = 9.57TIVDD59 pKa = 3.71TALSDD64 pKa = 4.05DD65 pKa = 4.41LVVLQYY71 pKa = 10.86RR72 pKa = 11.84YY73 pKa = 8.72TNDD76 pKa = 3.04GSVTSVTIYY85 pKa = 11.0YY86 pKa = 10.13NFAISRR92 pKa = 11.84LTTVEE97 pKa = 4.29TIAGEE102 pKa = 4.09NIYY105 pKa = 10.81RR106 pKa = 11.84RR107 pKa = 11.84DD108 pKa = 3.69TFLSDD113 pKa = 3.46YY114 pKa = 11.2NFGDD118 pKa = 3.55MVKK121 pKa = 10.28ISVVPANDD129 pKa = 5.29LITIYY134 pKa = 10.66INGVRR139 pKa = 11.84VYY141 pKa = 10.81GYY143 pKa = 7.59EE144 pKa = 3.89TEE146 pKa = 4.23LNKK149 pKa = 10.62GVASSTIKK157 pKa = 10.99LNGTAPVLTEE167 pKa = 5.15LEE169 pKa = 4.34YY170 pKa = 10.91TDD172 pKa = 4.82SNEE175 pKa = 4.0GNSDD179 pKa = 3.14IQSPLAALPLSLIGKK194 pKa = 7.34STQFQKK200 pKa = 11.02LGIMHH205 pKa = 7.09NKK207 pKa = 8.42PSTMSFVRR215 pKa = 11.84EE216 pKa = 4.46FWAEE220 pKa = 4.05TVDD223 pKa = 3.38TSMFPGWPTTRR234 pKa = 11.84YY235 pKa = 7.97PRR237 pKa = 11.84ARR239 pKa = 11.84FSCTDD244 pKa = 3.19HH245 pKa = 7.09DD246 pKa = 4.3TGSGGIWVRR255 pKa = 11.84VYY257 pKa = 11.14DD258 pKa = 4.34KK259 pKa = 11.27DD260 pKa = 3.88LGAITDD266 pKa = 3.85PSSWLEE272 pKa = 3.65WDD274 pKa = 4.22DD275 pKa = 3.59VSSRR279 pKa = 11.84PEE281 pKa = 3.46FDD283 pKa = 3.83HH284 pKa = 7.6IEE286 pKa = 4.06RR287 pKa = 11.84KK288 pKa = 9.25INPVFVYY295 pKa = 10.41SGTGDD300 pKa = 3.36QTEE303 pKa = 4.53TPMHH307 pKa = 6.52LVAADD312 pKa = 4.07GDD314 pKa = 3.71ILMYY318 pKa = 9.21FHH320 pKa = 7.27NSTVTLEE327 pKa = 4.14GFPAGIQATHH337 pKa = 6.43YY338 pKa = 7.9ATTLNGIDD346 pKa = 3.88YY347 pKa = 8.5TDD349 pKa = 4.1PTPSSIVYY357 pKa = 10.23DD358 pKa = 3.68PTYY361 pKa = 11.0LEE363 pKa = 4.91GDD365 pKa = 3.35GHH367 pKa = 6.21TGYY370 pKa = 10.11FQPGRR375 pKa = 11.84NTFAEE380 pKa = 4.13IPYY383 pKa = 9.68KK384 pKa = 10.76YY385 pKa = 9.92IGIHH389 pKa = 4.66LHH391 pKa = 6.17GGAGASRR398 pKa = 11.84GSCWAIEE405 pKa = 4.05VSNDD409 pKa = 2.73GKK411 pKa = 10.56NWEE414 pKa = 3.96RR415 pKa = 11.84WRR417 pKa = 11.84TFMRR421 pKa = 11.84DD422 pKa = 3.33FQATKK427 pKa = 10.18RR428 pKa = 11.84LPYY431 pKa = 9.83TDD433 pKa = 5.03EE434 pKa = 3.76EE435 pKa = 4.81DD436 pKa = 3.73KK437 pKa = 11.28FFVLQRR443 pKa = 11.84VHH445 pKa = 5.03EE446 pKa = 4.36AKK448 pKa = 10.39RR449 pKa = 11.84VGGYY453 pKa = 9.21YY454 pKa = 9.69RR455 pKa = 11.84VWGRR459 pKa = 11.84WQRR462 pKa = 11.84EE463 pKa = 4.02AFGGRR468 pKa = 11.84LGEE471 pKa = 4.15LEE473 pKa = 4.13AVEE476 pKa = 4.4VLVDD480 pKa = 3.47SDD482 pKa = 4.05FNIVSEE488 pKa = 4.71CNVFIPRR495 pKa = 11.84GSEE498 pKa = 4.1GEE500 pKa = 3.67FDD502 pKa = 3.34AVEE505 pKa = 3.65IRR507 pKa = 11.84GYY509 pKa = 10.95FPFEE513 pKa = 4.03YY514 pKa = 10.6GGVEE518 pKa = 3.89YY519 pKa = 8.81ATYY522 pKa = 8.97STRR525 pKa = 11.84LGTTIDD531 pKa = 3.97DD532 pKa = 3.89EE533 pKa = 4.62SAIGIGIVTDD543 pKa = 3.87VNRR546 pKa = 11.84NWQIYY551 pKa = 10.38NGMANKK557 pKa = 10.04SRR559 pKa = 11.84TLLVDD564 pKa = 3.71RR565 pKa = 11.84SNFPEE570 pKa = 4.25KK571 pKa = 10.54VSSSEE576 pKa = 4.16TIVLKK581 pKa = 10.28TEE583 pKa = 3.98AGYY586 pKa = 10.42QYY588 pKa = 10.75QSLPIRR594 pKa = 11.84RR595 pKa = 11.84DD596 pKa = 3.21NSISTLISNDD606 pKa = 3.36PVDD609 pKa = 4.84LSGQITDD616 pKa = 3.45VTFKK620 pKa = 11.35NIGKK624 pKa = 10.14DD625 pKa = 3.21SAAPIYY631 pKa = 10.73LEE633 pKa = 4.83FGITDD638 pKa = 3.68SLSNPTQQLSISFDD652 pKa = 3.43TDD654 pKa = 2.96HH655 pKa = 7.55DD656 pKa = 4.58VVTGCIPMLMRR667 pKa = 11.84IIGTVGTQEE676 pKa = 3.7VRR678 pKa = 11.84SINVFGMDD686 pKa = 3.35NDD688 pKa = 3.8NPSVYY693 pKa = 8.82EE694 pKa = 3.91TANTKK699 pKa = 10.35HH700 pKa = 6.32EE701 pKa = 3.99LTIRR705 pKa = 11.84IDD707 pKa = 3.14KK708 pKa = 10.42HH709 pKa = 6.55NNEE712 pKa = 4.04IQFLSGGSVTDD723 pKa = 3.6TLPLAGFVADD733 pKa = 3.42TPMFVFIKK741 pKa = 10.49AGTIDD746 pKa = 3.79PDD748 pKa = 3.49EE749 pKa = 4.91TEE751 pKa = 4.53DD752 pKa = 3.98SSVSFNEE759 pKa = 3.88ISVQSATADD768 pKa = 3.47KK769 pKa = 10.9LVEE772 pKa = 4.41LVNAHH777 pKa = 7.34PIADD781 pKa = 4.35AGPDD785 pKa = 3.34QSGISAGSTFQLDD798 pKa = 3.53GSGSYY803 pKa = 10.91DD804 pKa = 3.15SDD806 pKa = 3.98GSIVGYY812 pKa = 10.19EE813 pKa = 3.87WVQVDD818 pKa = 3.44NGAPSVAPFDD828 pKa = 3.97TTAVQPMVTAPSIGTPTTLRR848 pKa = 11.84FEE850 pKa = 4.81LVVEE854 pKa = 4.77DD855 pKa = 4.71DD856 pKa = 4.91DD857 pKa = 5.45GAFSVADD864 pKa = 3.62AVNVEE869 pKa = 4.53VTSTPNVGPTANAGTPSTIEE889 pKa = 4.37AGSQYY894 pKa = 10.74QLNGSASDD902 pKa = 3.47SDD904 pKa = 3.77GTIVSTVWTQTGDD917 pKa = 3.29AVTLSNPNVLNPTFTAPLVSSGSTTFTLTVTDD949 pKa = 4.99DD950 pKa = 4.29DD951 pKa = 5.55GEE953 pKa = 4.56TATDD957 pKa = 3.61TVVITYY963 pKa = 9.65EE964 pKa = 4.02DD965 pKa = 3.34TTAPVITRR973 pKa = 11.84NGPATVTLNLNDD985 pKa = 3.86TFTPATFTAFDD996 pKa = 4.2AVDD999 pKa = 3.88GDD1001 pKa = 4.07LTSAITVVNPYY1012 pKa = 10.14QPATGEE1018 pKa = 4.15YY1019 pKa = 8.73TVTANVSDD1027 pKa = 3.98AAGNAATEE1035 pKa = 4.37VTQVVTVTDD1044 pKa = 4.74DD1045 pKa = 2.63ISYY1048 pKa = 9.8PFNTVSSEE1056 pKa = 4.03NIPFGNNVVLQTTIRR1071 pKa = 11.84LGSTRR1076 pKa = 11.84CFNVNLSSWCNPEE1089 pKa = 3.73EE1090 pKa = 4.54LYY1092 pKa = 11.01SHH1094 pKa = 6.61QIDD1097 pKa = 3.73VSGTGLSIVNSQIQGGILVFYY1118 pKa = 10.52AQGLAVGRR1126 pKa = 11.84HH1127 pKa = 4.98KK1128 pKa = 10.91FKK1130 pKa = 9.82ITYY1133 pKa = 7.49STQSRR1138 pKa = 11.84QDD1140 pKa = 3.01YY1141 pKa = 8.8FYY1143 pKa = 10.95AYY1145 pKa = 10.51VEE1147 pKa = 4.21VSSYY1151 pKa = 11.36EE1152 pKa = 4.03GLL1154 pKa = 3.64
MM1 pKa = 7.72AVTTLDD7 pKa = 3.54LVQTNGTPLPSEE19 pKa = 4.05LTVLEE24 pKa = 4.74GEE26 pKa = 4.37VEE28 pKa = 4.4YY29 pKa = 11.33NGSGLVAATAPAALNFNHH47 pKa = 7.52KK48 pKa = 9.09DD49 pKa = 3.4TAIVSYY55 pKa = 9.57TIVDD59 pKa = 3.71TALSDD64 pKa = 4.05DD65 pKa = 4.41LVVLQYY71 pKa = 10.86RR72 pKa = 11.84YY73 pKa = 8.72TNDD76 pKa = 3.04GSVTSVTIYY85 pKa = 11.0YY86 pKa = 10.13NFAISRR92 pKa = 11.84LTTVEE97 pKa = 4.29TIAGEE102 pKa = 4.09NIYY105 pKa = 10.81RR106 pKa = 11.84RR107 pKa = 11.84DD108 pKa = 3.69TFLSDD113 pKa = 3.46YY114 pKa = 11.2NFGDD118 pKa = 3.55MVKK121 pKa = 10.28ISVVPANDD129 pKa = 5.29LITIYY134 pKa = 10.66INGVRR139 pKa = 11.84VYY141 pKa = 10.81GYY143 pKa = 7.59EE144 pKa = 3.89TEE146 pKa = 4.23LNKK149 pKa = 10.62GVASSTIKK157 pKa = 10.99LNGTAPVLTEE167 pKa = 5.15LEE169 pKa = 4.34YY170 pKa = 10.91TDD172 pKa = 4.82SNEE175 pKa = 4.0GNSDD179 pKa = 3.14IQSPLAALPLSLIGKK194 pKa = 7.34STQFQKK200 pKa = 11.02LGIMHH205 pKa = 7.09NKK207 pKa = 8.42PSTMSFVRR215 pKa = 11.84EE216 pKa = 4.46FWAEE220 pKa = 4.05TVDD223 pKa = 3.38TSMFPGWPTTRR234 pKa = 11.84YY235 pKa = 7.97PRR237 pKa = 11.84ARR239 pKa = 11.84FSCTDD244 pKa = 3.19HH245 pKa = 7.09DD246 pKa = 4.3TGSGGIWVRR255 pKa = 11.84VYY257 pKa = 11.14DD258 pKa = 4.34KK259 pKa = 11.27DD260 pKa = 3.88LGAITDD266 pKa = 3.85PSSWLEE272 pKa = 3.65WDD274 pKa = 4.22DD275 pKa = 3.59VSSRR279 pKa = 11.84PEE281 pKa = 3.46FDD283 pKa = 3.83HH284 pKa = 7.6IEE286 pKa = 4.06RR287 pKa = 11.84KK288 pKa = 9.25INPVFVYY295 pKa = 10.41SGTGDD300 pKa = 3.36QTEE303 pKa = 4.53TPMHH307 pKa = 6.52LVAADD312 pKa = 4.07GDD314 pKa = 3.71ILMYY318 pKa = 9.21FHH320 pKa = 7.27NSTVTLEE327 pKa = 4.14GFPAGIQATHH337 pKa = 6.43YY338 pKa = 7.9ATTLNGIDD346 pKa = 3.88YY347 pKa = 8.5TDD349 pKa = 4.1PTPSSIVYY357 pKa = 10.23DD358 pKa = 3.68PTYY361 pKa = 11.0LEE363 pKa = 4.91GDD365 pKa = 3.35GHH367 pKa = 6.21TGYY370 pKa = 10.11FQPGRR375 pKa = 11.84NTFAEE380 pKa = 4.13IPYY383 pKa = 9.68KK384 pKa = 10.76YY385 pKa = 9.92IGIHH389 pKa = 4.66LHH391 pKa = 6.17GGAGASRR398 pKa = 11.84GSCWAIEE405 pKa = 4.05VSNDD409 pKa = 2.73GKK411 pKa = 10.56NWEE414 pKa = 3.96RR415 pKa = 11.84WRR417 pKa = 11.84TFMRR421 pKa = 11.84DD422 pKa = 3.33FQATKK427 pKa = 10.18RR428 pKa = 11.84LPYY431 pKa = 9.83TDD433 pKa = 5.03EE434 pKa = 3.76EE435 pKa = 4.81DD436 pKa = 3.73KK437 pKa = 11.28FFVLQRR443 pKa = 11.84VHH445 pKa = 5.03EE446 pKa = 4.36AKK448 pKa = 10.39RR449 pKa = 11.84VGGYY453 pKa = 9.21YY454 pKa = 9.69RR455 pKa = 11.84VWGRR459 pKa = 11.84WQRR462 pKa = 11.84EE463 pKa = 4.02AFGGRR468 pKa = 11.84LGEE471 pKa = 4.15LEE473 pKa = 4.13AVEE476 pKa = 4.4VLVDD480 pKa = 3.47SDD482 pKa = 4.05FNIVSEE488 pKa = 4.71CNVFIPRR495 pKa = 11.84GSEE498 pKa = 4.1GEE500 pKa = 3.67FDD502 pKa = 3.34AVEE505 pKa = 3.65IRR507 pKa = 11.84GYY509 pKa = 10.95FPFEE513 pKa = 4.03YY514 pKa = 10.6GGVEE518 pKa = 3.89YY519 pKa = 8.81ATYY522 pKa = 8.97STRR525 pKa = 11.84LGTTIDD531 pKa = 3.97DD532 pKa = 3.89EE533 pKa = 4.62SAIGIGIVTDD543 pKa = 3.87VNRR546 pKa = 11.84NWQIYY551 pKa = 10.38NGMANKK557 pKa = 10.04SRR559 pKa = 11.84TLLVDD564 pKa = 3.71RR565 pKa = 11.84SNFPEE570 pKa = 4.25KK571 pKa = 10.54VSSSEE576 pKa = 4.16TIVLKK581 pKa = 10.28TEE583 pKa = 3.98AGYY586 pKa = 10.42QYY588 pKa = 10.75QSLPIRR594 pKa = 11.84RR595 pKa = 11.84DD596 pKa = 3.21NSISTLISNDD606 pKa = 3.36PVDD609 pKa = 4.84LSGQITDD616 pKa = 3.45VTFKK620 pKa = 11.35NIGKK624 pKa = 10.14DD625 pKa = 3.21SAAPIYY631 pKa = 10.73LEE633 pKa = 4.83FGITDD638 pKa = 3.68SLSNPTQQLSISFDD652 pKa = 3.43TDD654 pKa = 2.96HH655 pKa = 7.55DD656 pKa = 4.58VVTGCIPMLMRR667 pKa = 11.84IIGTVGTQEE676 pKa = 3.7VRR678 pKa = 11.84SINVFGMDD686 pKa = 3.35NDD688 pKa = 3.8NPSVYY693 pKa = 8.82EE694 pKa = 3.91TANTKK699 pKa = 10.35HH700 pKa = 6.32EE701 pKa = 3.99LTIRR705 pKa = 11.84IDD707 pKa = 3.14KK708 pKa = 10.42HH709 pKa = 6.55NNEE712 pKa = 4.04IQFLSGGSVTDD723 pKa = 3.6TLPLAGFVADD733 pKa = 3.42TPMFVFIKK741 pKa = 10.49AGTIDD746 pKa = 3.79PDD748 pKa = 3.49EE749 pKa = 4.91TEE751 pKa = 4.53DD752 pKa = 3.98SSVSFNEE759 pKa = 3.88ISVQSATADD768 pKa = 3.47KK769 pKa = 10.9LVEE772 pKa = 4.41LVNAHH777 pKa = 7.34PIADD781 pKa = 4.35AGPDD785 pKa = 3.34QSGISAGSTFQLDD798 pKa = 3.53GSGSYY803 pKa = 10.91DD804 pKa = 3.15SDD806 pKa = 3.98GSIVGYY812 pKa = 10.19EE813 pKa = 3.87WVQVDD818 pKa = 3.44NGAPSVAPFDD828 pKa = 3.97TTAVQPMVTAPSIGTPTTLRR848 pKa = 11.84FEE850 pKa = 4.81LVVEE854 pKa = 4.77DD855 pKa = 4.71DD856 pKa = 4.91DD857 pKa = 5.45GAFSVADD864 pKa = 3.62AVNVEE869 pKa = 4.53VTSTPNVGPTANAGTPSTIEE889 pKa = 4.37AGSQYY894 pKa = 10.74QLNGSASDD902 pKa = 3.47SDD904 pKa = 3.77GTIVSTVWTQTGDD917 pKa = 3.29AVTLSNPNVLNPTFTAPLVSSGSTTFTLTVTDD949 pKa = 4.99DD950 pKa = 4.29DD951 pKa = 5.55GEE953 pKa = 4.56TATDD957 pKa = 3.61TVVITYY963 pKa = 9.65EE964 pKa = 4.02DD965 pKa = 3.34TTAPVITRR973 pKa = 11.84NGPATVTLNLNDD985 pKa = 3.86TFTPATFTAFDD996 pKa = 4.2AVDD999 pKa = 3.88GDD1001 pKa = 4.07LTSAITVVNPYY1012 pKa = 10.14QPATGEE1018 pKa = 4.15YY1019 pKa = 8.73TVTANVSDD1027 pKa = 3.98AAGNAATEE1035 pKa = 4.37VTQVVTVTDD1044 pKa = 4.74DD1045 pKa = 2.63ISYY1048 pKa = 9.8PFNTVSSEE1056 pKa = 4.03NIPFGNNVVLQTTIRR1071 pKa = 11.84LGSTRR1076 pKa = 11.84CFNVNLSSWCNPEE1089 pKa = 3.73EE1090 pKa = 4.54LYY1092 pKa = 11.01SHH1094 pKa = 6.61QIDD1097 pKa = 3.73VSGTGLSIVNSQIQGGILVFYY1118 pKa = 10.52AQGLAVGRR1126 pKa = 11.84HH1127 pKa = 4.98KK1128 pKa = 10.91FKK1130 pKa = 9.82ITYY1133 pKa = 7.49STQSRR1138 pKa = 11.84QDD1140 pKa = 3.01YY1141 pKa = 8.8FYY1143 pKa = 10.95AYY1145 pKa = 10.51VEE1147 pKa = 4.21VSSYY1151 pKa = 11.36EE1152 pKa = 4.03GLL1154 pKa = 3.64
Molecular weight: 125.41 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6M3YR60|A0A6M3YR60_9CAUD Uncharacterized protein OS=Alteromonas phage vB_AcoS-R7M OX=2729541 GN=vBAcoSR7M_2 PE=4 SV=1
MM1 pKa = 7.61SKK3 pKa = 8.66TAQQRR8 pKa = 11.84FDD10 pKa = 3.08EE11 pKa = 4.8TYY13 pKa = 7.62ITGYY17 pKa = 10.21EE18 pKa = 3.89IQTIMDD24 pKa = 3.82VNRR27 pKa = 11.84STVLNARR34 pKa = 11.84KK35 pKa = 9.68RR36 pKa = 11.84GILPQPIQVPGSRR49 pKa = 11.84SFIWEE54 pKa = 3.82RR55 pKa = 11.84TIVQPYY61 pKa = 7.9IDD63 pKa = 3.65AWKK66 pKa = 10.11VALASRR72 pKa = 11.84RR73 pKa = 11.84RR74 pKa = 11.84EE75 pKa = 3.96LTGGVV80 pKa = 3.34
MM1 pKa = 7.61SKK3 pKa = 8.66TAQQRR8 pKa = 11.84FDD10 pKa = 3.08EE11 pKa = 4.8TYY13 pKa = 7.62ITGYY17 pKa = 10.21EE18 pKa = 3.89IQTIMDD24 pKa = 3.82VNRR27 pKa = 11.84STVLNARR34 pKa = 11.84KK35 pKa = 9.68RR36 pKa = 11.84GILPQPIQVPGSRR49 pKa = 11.84SFIWEE54 pKa = 3.82RR55 pKa = 11.84TIVQPYY61 pKa = 7.9IDD63 pKa = 3.65AWKK66 pKa = 10.11VALASRR72 pKa = 11.84RR73 pKa = 11.84RR74 pKa = 11.84EE75 pKa = 3.96LTGGVV80 pKa = 3.34
Molecular weight: 9.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
17771 |
31 |
1528 |
265.2 |
29.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.996 ± 0.498 | 1.109 ± 0.128 |
5.959 ± 0.187 | 7.231 ± 0.363 |
3.697 ± 0.14 | 7.282 ± 0.228 |
1.823 ± 0.159 | 5.638 ± 0.21 |
5.717 ± 0.398 | 8.047 ± 0.273 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.47 ± 0.124 | 4.997 ± 0.135 |
4.316 ± 0.211 | 3.962 ± 0.391 |
5.036 ± 0.199 | 6.387 ± 0.267 |
6.331 ± 0.413 | 6.702 ± 0.235 |
1.474 ± 0.138 | 3.826 ± 0.236 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
