
Lake Sarah-associated circular virus-26
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 8.1
Get precalculated fractions of proteins
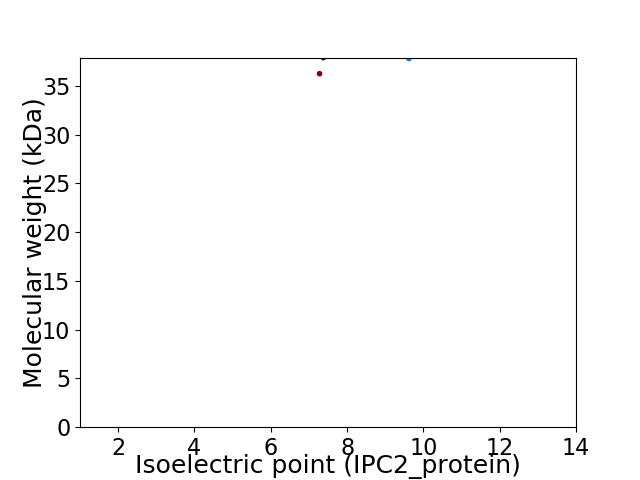
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
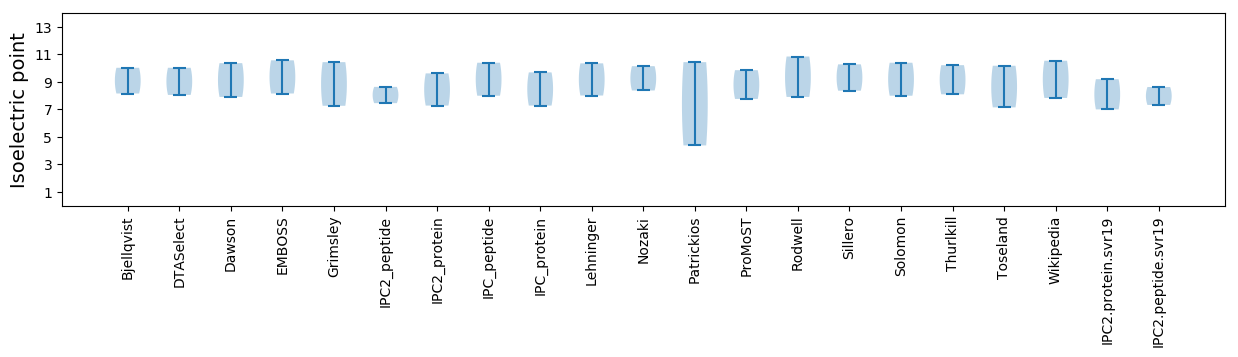
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A140AQM6|A0A140AQM6_9VIRU Putative capsid protein OS=Lake Sarah-associated circular virus-26 OX=1685753 PE=4 SV=1
MM1 pKa = 7.29VFNFNAKK8 pKa = 9.62HH9 pKa = 5.64VFLTYY14 pKa = 10.18PQTTLTKK21 pKa = 9.01EE22 pKa = 4.01ALYY25 pKa = 9.95EE26 pKa = 3.86ASIRR30 pKa = 11.84WGCVSGTIGQEE41 pKa = 3.54KK42 pKa = 9.11HH43 pKa = 5.53QDD45 pKa = 3.48GGLHH49 pKa = 5.76LHH51 pKa = 6.64AVFAFDD57 pKa = 3.6RR58 pKa = 11.84KK59 pKa = 9.47RR60 pKa = 11.84HH61 pKa = 5.26IRR63 pKa = 11.84DD64 pKa = 2.93EE65 pKa = 4.32RR66 pKa = 11.84FFDD69 pKa = 4.49LEE71 pKa = 4.61GYY73 pKa = 9.35HH74 pKa = 6.99PNVQSARR81 pKa = 11.84NVKK84 pKa = 9.82AVRR87 pKa = 11.84EE88 pKa = 4.14YY89 pKa = 11.14CEE91 pKa = 4.99KK92 pKa = 10.54EE93 pKa = 3.88DD94 pKa = 4.8PEE96 pKa = 4.12ALRR99 pKa = 11.84YY100 pKa = 9.26GAFEE104 pKa = 4.29GCEE107 pKa = 3.96KK108 pKa = 10.33IDD110 pKa = 3.47WKK112 pKa = 11.21DD113 pKa = 3.66LLGLATDD120 pKa = 3.65SAHH123 pKa = 7.15FIKK126 pKa = 10.7LVTDD130 pKa = 4.05RR131 pKa = 11.84DD132 pKa = 3.53VSFCVKK138 pKa = 10.13HH139 pKa = 5.77FSNVRR144 pKa = 11.84AFAEE148 pKa = 4.15YY149 pKa = 9.48TFRR152 pKa = 11.84SRR154 pKa = 11.84KK155 pKa = 9.28EE156 pKa = 3.75DD157 pKa = 3.43FTPQFTDD164 pKa = 3.43FRR166 pKa = 11.84NQLRR170 pKa = 11.84PMQDD174 pKa = 2.62FHH176 pKa = 9.58AEE178 pKa = 3.98LLRR181 pKa = 11.84PSRR184 pKa = 11.84PRR186 pKa = 11.84EE187 pKa = 3.53RR188 pKa = 11.84HH189 pKa = 3.95TSIVIVGATRR199 pKa = 11.84LGKK202 pKa = 8.09TEE204 pKa = 3.84WARR207 pKa = 11.84SLGNHH212 pKa = 6.34TYY214 pKa = 10.19WSGAVDD220 pKa = 3.41WTRR223 pKa = 11.84HH224 pKa = 5.6DD225 pKa = 3.29NHH227 pKa = 6.5SRR229 pKa = 11.84YY230 pKa = 10.08AVIDD234 pKa = 4.21DD235 pKa = 4.97LSLEE239 pKa = 4.3YY240 pKa = 10.01IPRR243 pKa = 11.84LQQIFGCQHH252 pKa = 6.6NISVSQKK259 pKa = 8.93YY260 pKa = 9.28KK261 pKa = 10.71PIMTFDD267 pKa = 2.79WGIPVIYY274 pKa = 8.99LTNDD278 pKa = 2.72EE279 pKa = 5.12NYY281 pKa = 10.67LEE283 pKa = 4.45KK284 pKa = 10.85QPDD287 pKa = 4.47FIKK290 pKa = 10.51SWWQNNVVTCHH301 pKa = 5.17VRR303 pKa = 11.84NKK305 pKa = 10.76LFVV308 pKa = 3.33
MM1 pKa = 7.29VFNFNAKK8 pKa = 9.62HH9 pKa = 5.64VFLTYY14 pKa = 10.18PQTTLTKK21 pKa = 9.01EE22 pKa = 4.01ALYY25 pKa = 9.95EE26 pKa = 3.86ASIRR30 pKa = 11.84WGCVSGTIGQEE41 pKa = 3.54KK42 pKa = 9.11HH43 pKa = 5.53QDD45 pKa = 3.48GGLHH49 pKa = 5.76LHH51 pKa = 6.64AVFAFDD57 pKa = 3.6RR58 pKa = 11.84KK59 pKa = 9.47RR60 pKa = 11.84HH61 pKa = 5.26IRR63 pKa = 11.84DD64 pKa = 2.93EE65 pKa = 4.32RR66 pKa = 11.84FFDD69 pKa = 4.49LEE71 pKa = 4.61GYY73 pKa = 9.35HH74 pKa = 6.99PNVQSARR81 pKa = 11.84NVKK84 pKa = 9.82AVRR87 pKa = 11.84EE88 pKa = 4.14YY89 pKa = 11.14CEE91 pKa = 4.99KK92 pKa = 10.54EE93 pKa = 3.88DD94 pKa = 4.8PEE96 pKa = 4.12ALRR99 pKa = 11.84YY100 pKa = 9.26GAFEE104 pKa = 4.29GCEE107 pKa = 3.96KK108 pKa = 10.33IDD110 pKa = 3.47WKK112 pKa = 11.21DD113 pKa = 3.66LLGLATDD120 pKa = 3.65SAHH123 pKa = 7.15FIKK126 pKa = 10.7LVTDD130 pKa = 4.05RR131 pKa = 11.84DD132 pKa = 3.53VSFCVKK138 pKa = 10.13HH139 pKa = 5.77FSNVRR144 pKa = 11.84AFAEE148 pKa = 4.15YY149 pKa = 9.48TFRR152 pKa = 11.84SRR154 pKa = 11.84KK155 pKa = 9.28EE156 pKa = 3.75DD157 pKa = 3.43FTPQFTDD164 pKa = 3.43FRR166 pKa = 11.84NQLRR170 pKa = 11.84PMQDD174 pKa = 2.62FHH176 pKa = 9.58AEE178 pKa = 3.98LLRR181 pKa = 11.84PSRR184 pKa = 11.84PRR186 pKa = 11.84EE187 pKa = 3.53RR188 pKa = 11.84HH189 pKa = 3.95TSIVIVGATRR199 pKa = 11.84LGKK202 pKa = 8.09TEE204 pKa = 3.84WARR207 pKa = 11.84SLGNHH212 pKa = 6.34TYY214 pKa = 10.19WSGAVDD220 pKa = 3.41WTRR223 pKa = 11.84HH224 pKa = 5.6DD225 pKa = 3.29NHH227 pKa = 6.5SRR229 pKa = 11.84YY230 pKa = 10.08AVIDD234 pKa = 4.21DD235 pKa = 4.97LSLEE239 pKa = 4.3YY240 pKa = 10.01IPRR243 pKa = 11.84LQQIFGCQHH252 pKa = 6.6NISVSQKK259 pKa = 8.93YY260 pKa = 9.28KK261 pKa = 10.71PIMTFDD267 pKa = 2.79WGIPVIYY274 pKa = 8.99LTNDD278 pKa = 2.72EE279 pKa = 5.12NYY281 pKa = 10.67LEE283 pKa = 4.45KK284 pKa = 10.85QPDD287 pKa = 4.47FIKK290 pKa = 10.51SWWQNNVVTCHH301 pKa = 5.17VRR303 pKa = 11.84NKK305 pKa = 10.76LFVV308 pKa = 3.33
Molecular weight: 36.26 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140AQM6|A0A140AQM6_9VIRU Putative capsid protein OS=Lake Sarah-associated circular virus-26 OX=1685753 PE=4 SV=1
MM1 pKa = 7.46VKK3 pKa = 10.05RR4 pKa = 11.84KK5 pKa = 10.05YY6 pKa = 10.56SATGYY11 pKa = 9.82ARR13 pKa = 11.84NYY15 pKa = 8.92GKK17 pKa = 10.0NVLTAARR24 pKa = 11.84TLMSLRR30 pKa = 11.84NGFKK34 pKa = 10.42SGSRR38 pKa = 11.84GPNSTNNRR46 pKa = 11.84RR47 pKa = 11.84SFNSTGITTDD57 pKa = 3.35HH58 pKa = 6.73YY59 pKa = 11.61DD60 pKa = 2.94KK61 pKa = 10.16KK62 pKa = 9.42TKK64 pKa = 10.19YY65 pKa = 9.35RR66 pKa = 11.84YY67 pKa = 10.14KK68 pKa = 10.5RR69 pKa = 11.84MPYY72 pKa = 9.65RR73 pKa = 11.84KK74 pKa = 8.23RR75 pKa = 11.84KK76 pKa = 9.33RR77 pKa = 11.84YY78 pKa = 8.42VRR80 pKa = 11.84RR81 pKa = 11.84LKK83 pKa = 10.31FVKK86 pKa = 10.16HH87 pKa = 5.99ALSKK91 pKa = 11.02DD92 pKa = 2.97LATFKK97 pKa = 8.53TTYY100 pKa = 8.97ATSTAVTALAGAQNVFATFSVMGVPVTGANLQTAGNFNQTINAWQNMSGTIDD152 pKa = 3.28ASTAGDD158 pKa = 3.52HH159 pKa = 6.87YY160 pKa = 11.19GEE162 pKa = 4.25KK163 pKa = 9.51WKK165 pKa = 10.54QLSSVVNVSITNYY178 pKa = 8.49GTTTVIMDD186 pKa = 3.75VYY188 pKa = 10.72RR189 pKa = 11.84VVCRR193 pKa = 11.84KK194 pKa = 7.77TFSNSDD200 pKa = 3.34FSSPEE205 pKa = 3.85TTNGSSEE212 pKa = 4.24TVLMKK217 pKa = 10.81KK218 pKa = 9.91MSDD221 pKa = 3.44NLSGTVSNDD230 pKa = 2.59WEE232 pKa = 4.82TVGITPFAINAFTKK246 pKa = 10.1HH247 pKa = 5.92FKK249 pKa = 10.06ILQVKK254 pKa = 8.38EE255 pKa = 3.96VQIPAGNTTTLSFRR269 pKa = 11.84DD270 pKa = 3.53PKK272 pKa = 10.93NYY274 pKa = 9.85SFNMSQFLGKK284 pKa = 10.5VFMKK288 pKa = 10.55GLTKK292 pKa = 10.49GYY294 pKa = 9.65VYY296 pKa = 10.62RR297 pKa = 11.84FRR299 pKa = 11.84AIAATAGGEE308 pKa = 4.13SAAISMGVHH317 pKa = 5.87EE318 pKa = 5.41EE319 pKa = 3.96YY320 pKa = 11.38SNIVQEE326 pKa = 4.4LEE328 pKa = 3.96PTEE331 pKa = 4.12SLNYY335 pKa = 9.51HH336 pKa = 6.11ANKK339 pKa = 8.74TT340 pKa = 3.73
MM1 pKa = 7.46VKK3 pKa = 10.05RR4 pKa = 11.84KK5 pKa = 10.05YY6 pKa = 10.56SATGYY11 pKa = 9.82ARR13 pKa = 11.84NYY15 pKa = 8.92GKK17 pKa = 10.0NVLTAARR24 pKa = 11.84TLMSLRR30 pKa = 11.84NGFKK34 pKa = 10.42SGSRR38 pKa = 11.84GPNSTNNRR46 pKa = 11.84RR47 pKa = 11.84SFNSTGITTDD57 pKa = 3.35HH58 pKa = 6.73YY59 pKa = 11.61DD60 pKa = 2.94KK61 pKa = 10.16KK62 pKa = 9.42TKK64 pKa = 10.19YY65 pKa = 9.35RR66 pKa = 11.84YY67 pKa = 10.14KK68 pKa = 10.5RR69 pKa = 11.84MPYY72 pKa = 9.65RR73 pKa = 11.84KK74 pKa = 8.23RR75 pKa = 11.84KK76 pKa = 9.33RR77 pKa = 11.84YY78 pKa = 8.42VRR80 pKa = 11.84RR81 pKa = 11.84LKK83 pKa = 10.31FVKK86 pKa = 10.16HH87 pKa = 5.99ALSKK91 pKa = 11.02DD92 pKa = 2.97LATFKK97 pKa = 8.53TTYY100 pKa = 8.97ATSTAVTALAGAQNVFATFSVMGVPVTGANLQTAGNFNQTINAWQNMSGTIDD152 pKa = 3.28ASTAGDD158 pKa = 3.52HH159 pKa = 6.87YY160 pKa = 11.19GEE162 pKa = 4.25KK163 pKa = 9.51WKK165 pKa = 10.54QLSSVVNVSITNYY178 pKa = 8.49GTTTVIMDD186 pKa = 3.75VYY188 pKa = 10.72RR189 pKa = 11.84VVCRR193 pKa = 11.84KK194 pKa = 7.77TFSNSDD200 pKa = 3.34FSSPEE205 pKa = 3.85TTNGSSEE212 pKa = 4.24TVLMKK217 pKa = 10.81KK218 pKa = 9.91MSDD221 pKa = 3.44NLSGTVSNDD230 pKa = 2.59WEE232 pKa = 4.82TVGITPFAINAFTKK246 pKa = 10.1HH247 pKa = 5.92FKK249 pKa = 10.06ILQVKK254 pKa = 8.38EE255 pKa = 3.96VQIPAGNTTTLSFRR269 pKa = 11.84DD270 pKa = 3.53PKK272 pKa = 10.93NYY274 pKa = 9.85SFNMSQFLGKK284 pKa = 10.5VFMKK288 pKa = 10.55GLTKK292 pKa = 10.49GYY294 pKa = 9.65VYY296 pKa = 10.62RR297 pKa = 11.84FRR299 pKa = 11.84AIAATAGGEE308 pKa = 4.13SAAISMGVHH317 pKa = 5.87EE318 pKa = 5.41EE319 pKa = 3.96YY320 pKa = 11.38SNIVQEE326 pKa = 4.4LEE328 pKa = 3.96PTEE331 pKa = 4.12SLNYY335 pKa = 9.51HH336 pKa = 6.11ANKK339 pKa = 8.74TT340 pKa = 3.73
Molecular weight: 37.81 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
648 |
308 |
340 |
324.0 |
37.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.944 ± 0.75 | 1.08 ± 0.592 |
4.63 ± 1.271 | 4.63 ± 1.049 |
5.864 ± 0.65 | 6.019 ± 0.783 |
3.241 ± 1.111 | 4.167 ± 0.48 |
6.79 ± 0.866 | 6.019 ± 0.767 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.16 ± 0.809 | 6.173 ± 1.11 |
2.932 ± 0.436 | 3.395 ± 0.563 |
6.636 ± 0.788 | 7.253 ± 1.403 |
8.796 ± 2.013 | 7.099 ± 0.191 |
1.698 ± 0.614 | 4.475 ± 0.395 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
