
Bacilladnavirus sp.
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Baphyvirales; Bacilladnaviridae; Bacilladnavirus; unclassified Bacilladnavirus
Average proteome isoelectric point is 7.32
Get precalculated fractions of proteins
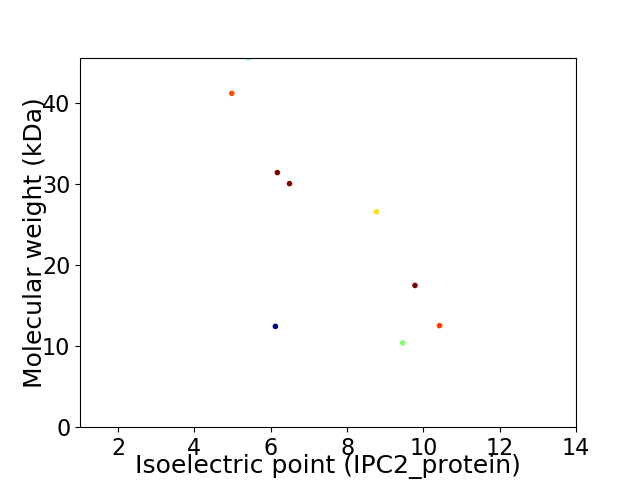
Virtual 2D-PAGE plot for 9 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A345N1F1|A0A345N1F1_9VIRU Uncharacterized protein OS=Bacilladnavirus sp. OX=2267680 PE=4 SV=1
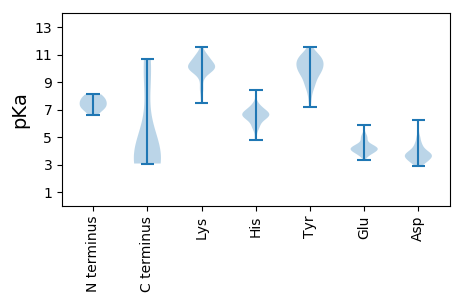
MM1 pKa = 7.09QHH3 pKa = 6.32LQFFAQTHH11 pKa = 6.67LYY13 pKa = 9.86CSPATFLHH21 pKa = 6.61SGVQFPWRR29 pKa = 11.84STIPFGTTAKK39 pKa = 10.14QSRR42 pKa = 11.84ADD44 pKa = 3.52MTRR47 pKa = 11.84CCCCPNNEE55 pKa = 4.09QTTGYY60 pKa = 10.26APPAFNPSPTGPNVPGIGDD79 pKa = 4.26DD80 pKa = 3.39IFGEE84 pKa = 4.14LHH86 pKa = 6.74SGIPEE91 pKa = 3.95NGLFPEE97 pKa = 5.0APCVKK102 pKa = 10.69NEE104 pKa = 4.04DD105 pKa = 4.23AYY107 pKa = 11.16DD108 pKa = 3.88DD109 pKa = 4.9EE110 pKa = 5.79INEE113 pKa = 4.56GGDD116 pKa = 3.45PSTAPVPPPVFTTQYY131 pKa = 9.84SPSFSAISGKK141 pKa = 10.63SSGDD145 pKa = 3.56CIQANTALYY154 pKa = 10.26ARR156 pKa = 11.84HH157 pKa = 6.73DD158 pKa = 3.93PDD160 pKa = 5.64GISGVTWSTWDD171 pKa = 3.74GITRR175 pKa = 11.84FNPCALTKK183 pKa = 10.22AQLYY187 pKa = 8.87AAIWPTGDD195 pKa = 3.52CRR197 pKa = 11.84NMLGLRR203 pKa = 11.84DD204 pKa = 4.51LYY206 pKa = 11.58YY207 pKa = 10.61SINPFADD214 pKa = 3.2VANPTIAEE222 pKa = 3.64IDD224 pKa = 3.14AWHH227 pKa = 6.98AIVVNHH233 pKa = 5.7YY234 pKa = 10.58RR235 pKa = 11.84DD236 pKa = 3.72LLGIATPILNDD247 pKa = 2.97ANLYY251 pKa = 9.87VRR253 pKa = 11.84AQWASEE259 pKa = 4.04RR260 pKa = 11.84KK261 pKa = 8.06NTTYY265 pKa = 10.79WDD267 pKa = 3.33TSYY270 pKa = 8.47PTGEE274 pKa = 4.15LCPTGSSTHH283 pKa = 6.47CGWTFIPSCLDD294 pKa = 3.14QAPYY298 pKa = 10.07LAPGQGCVASTPQSEE313 pKa = 5.25GIFGALPNPWSVRR326 pKa = 11.84FACVLAKK333 pKa = 10.11IVQTEE338 pKa = 4.88GITGHH343 pKa = 6.77GGPFLGRR350 pKa = 11.84PNMGMNFWCNGSEE363 pKa = 3.82TSLRR367 pKa = 11.84IKK369 pKa = 10.84FNGVPVNPCPPVV381 pKa = 3.07
MM1 pKa = 7.09QHH3 pKa = 6.32LQFFAQTHH11 pKa = 6.67LYY13 pKa = 9.86CSPATFLHH21 pKa = 6.61SGVQFPWRR29 pKa = 11.84STIPFGTTAKK39 pKa = 10.14QSRR42 pKa = 11.84ADD44 pKa = 3.52MTRR47 pKa = 11.84CCCCPNNEE55 pKa = 4.09QTTGYY60 pKa = 10.26APPAFNPSPTGPNVPGIGDD79 pKa = 4.26DD80 pKa = 3.39IFGEE84 pKa = 4.14LHH86 pKa = 6.74SGIPEE91 pKa = 3.95NGLFPEE97 pKa = 5.0APCVKK102 pKa = 10.69NEE104 pKa = 4.04DD105 pKa = 4.23AYY107 pKa = 11.16DD108 pKa = 3.88DD109 pKa = 4.9EE110 pKa = 5.79INEE113 pKa = 4.56GGDD116 pKa = 3.45PSTAPVPPPVFTTQYY131 pKa = 9.84SPSFSAISGKK141 pKa = 10.63SSGDD145 pKa = 3.56CIQANTALYY154 pKa = 10.26ARR156 pKa = 11.84HH157 pKa = 6.73DD158 pKa = 3.93PDD160 pKa = 5.64GISGVTWSTWDD171 pKa = 3.74GITRR175 pKa = 11.84FNPCALTKK183 pKa = 10.22AQLYY187 pKa = 8.87AAIWPTGDD195 pKa = 3.52CRR197 pKa = 11.84NMLGLRR203 pKa = 11.84DD204 pKa = 4.51LYY206 pKa = 11.58YY207 pKa = 10.61SINPFADD214 pKa = 3.2VANPTIAEE222 pKa = 3.64IDD224 pKa = 3.14AWHH227 pKa = 6.98AIVVNHH233 pKa = 5.7YY234 pKa = 10.58RR235 pKa = 11.84DD236 pKa = 3.72LLGIATPILNDD247 pKa = 2.97ANLYY251 pKa = 9.87VRR253 pKa = 11.84AQWASEE259 pKa = 4.04RR260 pKa = 11.84KK261 pKa = 8.06NTTYY265 pKa = 10.79WDD267 pKa = 3.33TSYY270 pKa = 8.47PTGEE274 pKa = 4.15LCPTGSSTHH283 pKa = 6.47CGWTFIPSCLDD294 pKa = 3.14QAPYY298 pKa = 10.07LAPGQGCVASTPQSEE313 pKa = 5.25GIFGALPNPWSVRR326 pKa = 11.84FACVLAKK333 pKa = 10.11IVQTEE338 pKa = 4.88GITGHH343 pKa = 6.77GGPFLGRR350 pKa = 11.84PNMGMNFWCNGSEE363 pKa = 3.82TSLRR367 pKa = 11.84IKK369 pKa = 10.84FNGVPVNPCPPVV381 pKa = 3.07
Molecular weight: 41.24 kDa
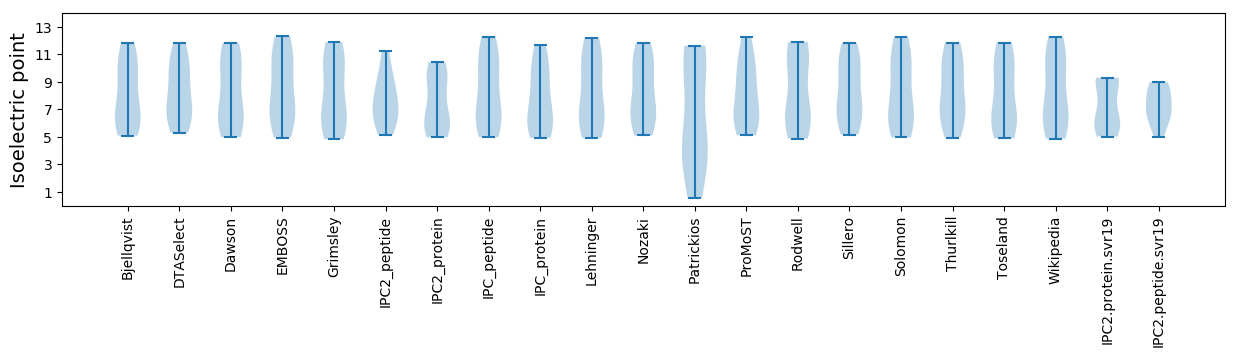
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A345N1E7|A0A345N1E7_9VIRU Uncharacterized protein OS=Bacilladnavirus sp. OX=2267680 PE=4 SV=1
MM1 pKa = 7.98KK2 pKa = 10.34IFDD5 pKa = 5.12DD6 pKa = 6.21DD7 pKa = 3.63NDD9 pKa = 3.28RR10 pKa = 11.84WITIGRR16 pKa = 11.84KK17 pKa = 8.47PRR19 pKa = 11.84YY20 pKa = 9.02NPRR23 pKa = 11.84TIANDD28 pKa = 3.86RR29 pKa = 11.84QWCSYY34 pKa = 9.78HH35 pKa = 7.22RR36 pKa = 11.84LPEE39 pKa = 3.83YY40 pKa = 10.51RR41 pKa = 11.84GLKK44 pKa = 9.77GFKK47 pKa = 9.67KK48 pKa = 10.3GLKK51 pKa = 9.32ARR53 pKa = 11.84QEE55 pKa = 4.0IFTTFQKK62 pKa = 10.37KK63 pKa = 10.2CSVALRR69 pKa = 11.84KK70 pKa = 10.09VYY72 pKa = 9.11EE73 pKa = 4.37AKK75 pKa = 10.4RR76 pKa = 11.84SMRR79 pKa = 11.84RR80 pKa = 11.84LLMNRR85 pKa = 11.84DD86 pKa = 3.88KK87 pKa = 11.52VCLDD91 pKa = 3.39ASIKK95 pKa = 9.58KK96 pKa = 9.54AAYY99 pKa = 9.25RR100 pKa = 11.84DD101 pKa = 3.6MYY103 pKa = 9.66RR104 pKa = 11.84WKK106 pKa = 10.53AIKK109 pKa = 9.62QDD111 pKa = 3.85RR112 pKa = 11.84LRR114 pKa = 11.84QAAKK118 pKa = 10.17LNAFRR123 pKa = 11.84RR124 pKa = 11.84SDD126 pKa = 3.47PAGYY130 pKa = 10.99AFMKK134 pKa = 10.66SFTPKK139 pKa = 10.09KK140 pKa = 10.04KK141 pKa = 9.92KK142 pKa = 10.26CKK144 pKa = 10.1CSS146 pKa = 3.21
MM1 pKa = 7.98KK2 pKa = 10.34IFDD5 pKa = 5.12DD6 pKa = 6.21DD7 pKa = 3.63NDD9 pKa = 3.28RR10 pKa = 11.84WITIGRR16 pKa = 11.84KK17 pKa = 8.47PRR19 pKa = 11.84YY20 pKa = 9.02NPRR23 pKa = 11.84TIANDD28 pKa = 3.86RR29 pKa = 11.84QWCSYY34 pKa = 9.78HH35 pKa = 7.22RR36 pKa = 11.84LPEE39 pKa = 3.83YY40 pKa = 10.51RR41 pKa = 11.84GLKK44 pKa = 9.77GFKK47 pKa = 9.67KK48 pKa = 10.3GLKK51 pKa = 9.32ARR53 pKa = 11.84QEE55 pKa = 4.0IFTTFQKK62 pKa = 10.37KK63 pKa = 10.2CSVALRR69 pKa = 11.84KK70 pKa = 10.09VYY72 pKa = 9.11EE73 pKa = 4.37AKK75 pKa = 10.4RR76 pKa = 11.84SMRR79 pKa = 11.84RR80 pKa = 11.84LLMNRR85 pKa = 11.84DD86 pKa = 3.88KK87 pKa = 11.52VCLDD91 pKa = 3.39ASIKK95 pKa = 9.58KK96 pKa = 9.54AAYY99 pKa = 9.25RR100 pKa = 11.84DD101 pKa = 3.6MYY103 pKa = 9.66RR104 pKa = 11.84WKK106 pKa = 10.53AIKK109 pKa = 9.62QDD111 pKa = 3.85RR112 pKa = 11.84LRR114 pKa = 11.84QAAKK118 pKa = 10.17LNAFRR123 pKa = 11.84RR124 pKa = 11.84SDD126 pKa = 3.47PAGYY130 pKa = 10.99AFMKK134 pKa = 10.66SFTPKK139 pKa = 10.09KK140 pKa = 10.04KK141 pKa = 9.92KK142 pKa = 10.26CKK144 pKa = 10.1CSS146 pKa = 3.21
Molecular weight: 17.51 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2022 |
84 |
404 |
224.7 |
25.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.578 ± 0.595 | 2.671 ± 0.778 |
5.885 ± 0.835 | 4.649 ± 0.602 |
4.303 ± 0.631 | 5.786 ± 0.882 |
2.226 ± 0.477 | 5.045 ± 0.496 |
6.33 ± 1.274 | 7.616 ± 0.968 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.978 ± 0.28 | 4.055 ± 0.686 |
8.111 ± 1.202 | 3.808 ± 0.198 |
6.38 ± 1.081 | 7.567 ± 0.966 |
6.034 ± 0.753 | 5.589 ± 0.925 |
1.632 ± 0.341 | 3.759 ± 0.469 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
