
Squirrel monkey polyomavirus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; Betapolyomavirus; Saimiri boliviensis polyomavirus 1
Average proteome isoelectric point is 6.52
Get precalculated fractions of proteins
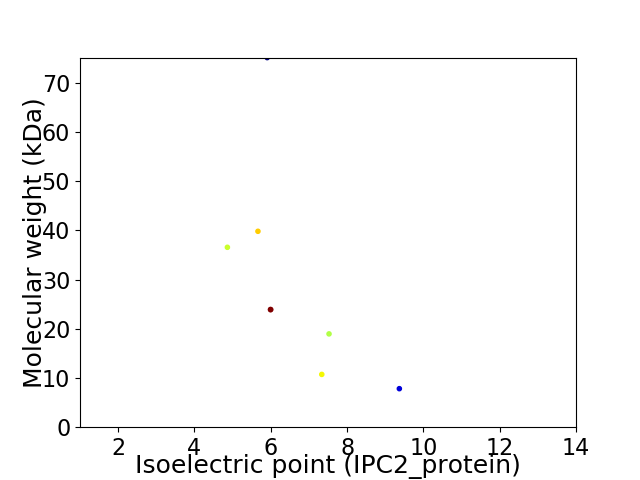
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
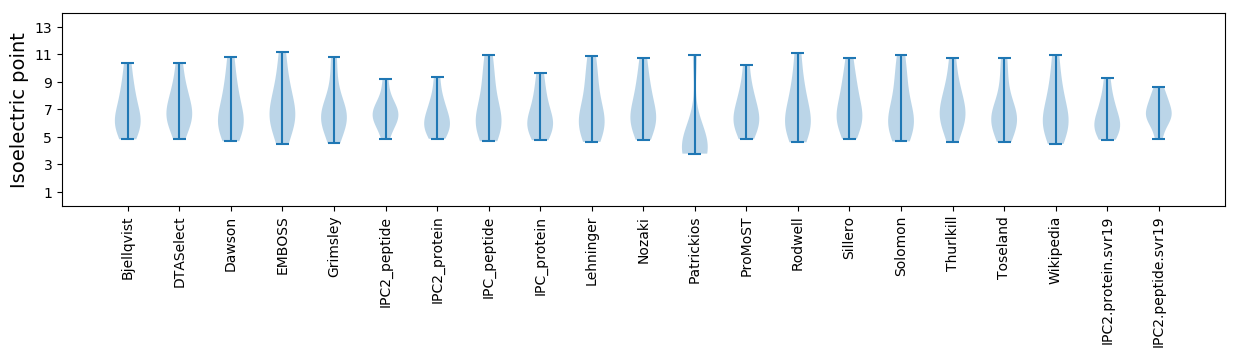
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|A8Y987-2|VP2-2_POVSM Isoform of A8Y987 Isoform VP3 of Minor capsid protein VP2 OS=Squirrel monkey polyomavirus OX=452475 GN=VP2 PE=3 SV=1
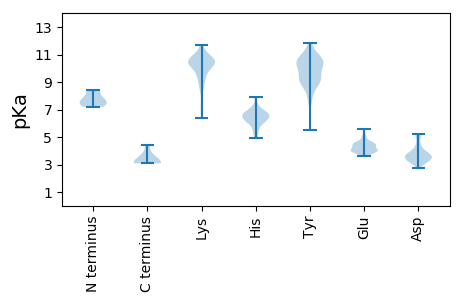
MM1 pKa = 7.31GALLAVLAEE10 pKa = 4.29VVEE13 pKa = 4.39LASVTGLSVEE23 pKa = 4.31SFISGEE29 pKa = 3.99AFATAEE35 pKa = 4.19LLEE38 pKa = 3.97AHH40 pKa = 7.15IANLVTVGGLTEE52 pKa = 4.49AEE54 pKa = 3.83ALAATEE60 pKa = 4.53VPAEE64 pKa = 4.29AYY66 pKa = 9.12AALTSLSSTFPQAFTAVAATEE87 pKa = 4.01LATTGTLTVGATVAAALYY105 pKa = 8.79PYY107 pKa = 9.81YY108 pKa = 10.37YY109 pKa = 10.45DD110 pKa = 3.48YY111 pKa = 10.56STPVANLNRR120 pKa = 11.84GLNPEE125 pKa = 4.22MALQLWFPEE134 pKa = 3.59IDD136 pKa = 3.62YY137 pKa = 9.26EE138 pKa = 5.16FPGLMPFVRR147 pKa = 11.84FINYY151 pKa = 9.15IDD153 pKa = 3.59PTQWATNLFEE163 pKa = 4.7TIGRR167 pKa = 11.84YY168 pKa = 7.68SWEE171 pKa = 3.68SAQRR175 pKa = 11.84YY176 pKa = 5.49GQNLIAHH183 pKa = 6.71EE184 pKa = 4.3VRR186 pKa = 11.84SASRR190 pKa = 11.84EE191 pKa = 3.93LATRR195 pKa = 11.84TAQGFSEE202 pKa = 4.24AVARR206 pKa = 11.84YY207 pKa = 9.01FEE209 pKa = 4.05NARR212 pKa = 11.84WAVSTLPRR220 pKa = 11.84SLYY223 pKa = 10.8SGLQSYY229 pKa = 9.15YY230 pKa = 10.58EE231 pKa = 4.12QLPSLNPMQVRR242 pKa = 11.84DD243 pKa = 3.22LHH245 pKa = 6.68RR246 pKa = 11.84RR247 pKa = 11.84LGQPIPNRR255 pKa = 11.84IALEE259 pKa = 3.91EE260 pKa = 3.87QAIKK264 pKa = 10.32SAEE267 pKa = 4.08YY268 pKa = 8.94VQKK271 pKa = 10.7VDD273 pKa = 3.87PPGGANQRR281 pKa = 11.84IAPDD285 pKa = 3.16WLLPLILGLYY295 pKa = 10.37GDD297 pKa = 5.2ISPSWEE303 pKa = 3.98STLEE307 pKa = 4.53DD308 pKa = 3.47IEE310 pKa = 4.55EE311 pKa = 4.46EE312 pKa = 3.8EE313 pKa = 5.56DD314 pKa = 3.62APQKK318 pKa = 10.58KK319 pKa = 9.27KK320 pKa = 10.74RR321 pKa = 11.84KK322 pKa = 6.86RR323 pKa = 11.84TKK325 pKa = 10.5KK326 pKa = 8.54NTPRR330 pKa = 11.84SAA332 pKa = 3.11
MM1 pKa = 7.31GALLAVLAEE10 pKa = 4.29VVEE13 pKa = 4.39LASVTGLSVEE23 pKa = 4.31SFISGEE29 pKa = 3.99AFATAEE35 pKa = 4.19LLEE38 pKa = 3.97AHH40 pKa = 7.15IANLVTVGGLTEE52 pKa = 4.49AEE54 pKa = 3.83ALAATEE60 pKa = 4.53VPAEE64 pKa = 4.29AYY66 pKa = 9.12AALTSLSSTFPQAFTAVAATEE87 pKa = 4.01LATTGTLTVGATVAAALYY105 pKa = 8.79PYY107 pKa = 9.81YY108 pKa = 10.37YY109 pKa = 10.45DD110 pKa = 3.48YY111 pKa = 10.56STPVANLNRR120 pKa = 11.84GLNPEE125 pKa = 4.22MALQLWFPEE134 pKa = 3.59IDD136 pKa = 3.62YY137 pKa = 9.26EE138 pKa = 5.16FPGLMPFVRR147 pKa = 11.84FINYY151 pKa = 9.15IDD153 pKa = 3.59PTQWATNLFEE163 pKa = 4.7TIGRR167 pKa = 11.84YY168 pKa = 7.68SWEE171 pKa = 3.68SAQRR175 pKa = 11.84YY176 pKa = 5.49GQNLIAHH183 pKa = 6.71EE184 pKa = 4.3VRR186 pKa = 11.84SASRR190 pKa = 11.84EE191 pKa = 3.93LATRR195 pKa = 11.84TAQGFSEE202 pKa = 4.24AVARR206 pKa = 11.84YY207 pKa = 9.01FEE209 pKa = 4.05NARR212 pKa = 11.84WAVSTLPRR220 pKa = 11.84SLYY223 pKa = 10.8SGLQSYY229 pKa = 9.15YY230 pKa = 10.58EE231 pKa = 4.12QLPSLNPMQVRR242 pKa = 11.84DD243 pKa = 3.22LHH245 pKa = 6.68RR246 pKa = 11.84RR247 pKa = 11.84LGQPIPNRR255 pKa = 11.84IALEE259 pKa = 3.91EE260 pKa = 3.87QAIKK264 pKa = 10.32SAEE267 pKa = 4.08YY268 pKa = 8.94VQKK271 pKa = 10.7VDD273 pKa = 3.87PPGGANQRR281 pKa = 11.84IAPDD285 pKa = 3.16WLLPLILGLYY295 pKa = 10.37GDD297 pKa = 5.2ISPSWEE303 pKa = 3.98STLEE307 pKa = 4.53DD308 pKa = 3.47IEE310 pKa = 4.55EE311 pKa = 4.46EE312 pKa = 3.8EE313 pKa = 5.56DD314 pKa = 3.62APQKK318 pKa = 10.58KK319 pKa = 9.27KK320 pKa = 10.74RR321 pKa = 11.84KK322 pKa = 6.86RR323 pKa = 11.84TKK325 pKa = 10.5KK326 pKa = 8.54NTPRR330 pKa = 11.84SAA332 pKa = 3.11
Molecular weight: 36.56 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|A8Y987|VP2_POVSM Minor capsid protein VP2 OS=Squirrel monkey polyomavirus OX=452475 GN=VP2 PE=3 SV=1
MM1 pKa = 7.37ARR3 pKa = 11.84KK4 pKa = 9.87LMARR8 pKa = 11.84KK9 pKa = 7.39TPPLVGLDD17 pKa = 3.6TVQLRR22 pKa = 11.84VEE24 pKa = 4.68TIWLTSQMDD33 pKa = 4.24LFVLTDD39 pKa = 3.25KK40 pKa = 11.47GEE42 pKa = 4.08PRR44 pKa = 11.84ITASRR49 pKa = 11.84ASASRR54 pKa = 11.84VATFGMTSVWLKK66 pKa = 10.57NSCFF70 pKa = 3.6
MM1 pKa = 7.37ARR3 pKa = 11.84KK4 pKa = 9.87LMARR8 pKa = 11.84KK9 pKa = 7.39TPPLVGLDD17 pKa = 3.6TVQLRR22 pKa = 11.84VEE24 pKa = 4.68TIWLTSQMDD33 pKa = 4.24LFVLTDD39 pKa = 3.25KK40 pKa = 11.47GEE42 pKa = 4.08PRR44 pKa = 11.84ITASRR49 pKa = 11.84ASASRR54 pKa = 11.84VATFGMTSVWLKK66 pKa = 10.57NSCFF70 pKa = 3.6
Molecular weight: 7.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
2085 |
70 |
655 |
260.6 |
29.6 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.619 ± 1.376 | 1.871 ± 0.685 |
4.365 ± 0.289 | 7.77 ± 0.36 |
4.221 ± 0.671 | 5.228 ± 0.455 |
1.439 ± 0.296 | 4.892 ± 0.329 |
6.811 ± 1.067 | 11.079 ± 0.653 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.158 ± 0.271 | 4.556 ± 0.551 |
5.851 ± 0.85 | 4.748 ± 0.326 |
4.604 ± 1.013 | 6.427 ± 0.354 |
6.187 ± 0.489 | 5.659 ± 0.643 |
1.727 ± 0.322 | 3.789 ± 0.349 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
