
Beauveria bassiana RNA virus 1
Taxonomy: Viruses; Riboviria; dsRNA viruses; unclassified dsRNA viruses
Average proteome isoelectric point is 6.06
Get precalculated fractions of proteins
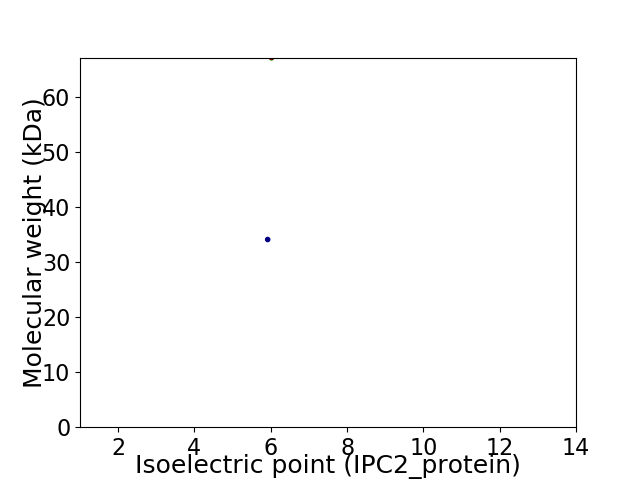
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0G4DJ74|A0A0G4DJ74_9VIRU Uncharacterized protein OS=Beauveria bassiana RNA virus 1 OX=1054649 PE=4 SV=1
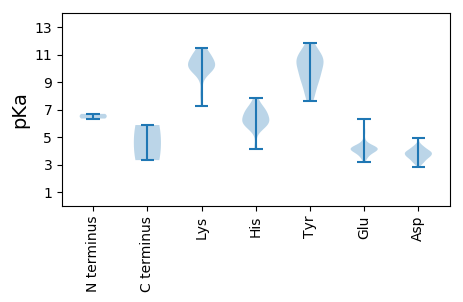
MM1 pKa = 6.68MVDD4 pKa = 4.2WIAEE8 pKa = 4.13NPSTVSEE15 pKa = 4.04ASGNGVKK22 pKa = 9.24EE23 pKa = 3.44WRR25 pKa = 11.84FFLKK29 pKa = 9.91HH30 pKa = 6.23CSWQRR35 pKa = 11.84RR36 pKa = 11.84LPGVSSLGHH45 pKa = 5.73SPAVRR50 pKa = 11.84YY51 pKa = 9.85RR52 pKa = 11.84YY53 pKa = 8.14VQPEE57 pKa = 4.15TSPSFLFSKK66 pKa = 10.54FLDD69 pKa = 3.89EE70 pKa = 4.55SQSSLGGIEE79 pKa = 4.24GPARR83 pKa = 11.84DD84 pKa = 3.65VLFSEE89 pKa = 5.33FVPPSEE95 pKa = 5.5DD96 pKa = 4.26DD97 pKa = 3.81LFEE100 pKa = 5.86HH101 pKa = 6.89IAGFGEE107 pKa = 4.29PGPDD111 pKa = 3.6GSHH114 pKa = 7.08LPGSDD119 pKa = 3.16FAQLFRR125 pKa = 11.84EE126 pKa = 4.41LRR128 pKa = 11.84RR129 pKa = 11.84LEE131 pKa = 3.97QWDD134 pKa = 3.97VPISGLLDD142 pKa = 3.07ATMLPKK148 pKa = 10.01IRR150 pKa = 11.84VPSATSPGIRR160 pKa = 11.84WKK162 pKa = 10.87KK163 pKa = 10.21LGYY166 pKa = 7.66KK167 pKa = 8.29TKK169 pKa = 10.53RR170 pKa = 11.84QALVPAVVEE179 pKa = 4.07ATRR182 pKa = 11.84ALNRR186 pKa = 11.84MVEE189 pKa = 4.19EE190 pKa = 4.21EE191 pKa = 4.1VAYY194 pKa = 8.04TVPPCGVAGRR204 pKa = 11.84GKK206 pKa = 10.09RR207 pKa = 11.84VNILEE212 pKa = 4.3GGSRR216 pKa = 11.84KK217 pKa = 9.63GKK219 pKa = 9.89KK220 pKa = 9.68DD221 pKa = 2.82GRR223 pKa = 11.84LIVMPDD229 pKa = 3.6LVRR232 pKa = 11.84HH233 pKa = 6.01LLGSLGSAPYY243 pKa = 9.48MSRR246 pKa = 11.84LRR248 pKa = 11.84GMDD251 pKa = 3.19HH252 pKa = 6.84SGGGVLLGMGPFQDD266 pKa = 3.9QYY268 pKa = 11.86QLIADD273 pKa = 4.41WATGASAFCFIDD285 pKa = 4.23FKK287 pKa = 11.43KK288 pKa = 10.49FDD290 pKa = 3.4QRR292 pKa = 11.84VPRR295 pKa = 11.84RR296 pKa = 11.84LLRR299 pKa = 11.84GVMNHH304 pKa = 5.77IRR306 pKa = 11.84SAFIGEE312 pKa = 4.25PGSRR316 pKa = 11.84AYY318 pKa = 9.44WEE320 pKa = 4.21SEE322 pKa = 3.48FRR324 pKa = 11.84HH325 pKa = 6.12LVDD328 pKa = 4.52TEE330 pKa = 3.66IAMPDD335 pKa = 3.75GSCYY339 pKa = 10.27RR340 pKa = 11.84KK341 pKa = 9.89RR342 pKa = 11.84GCVASGDD349 pKa = 3.84PWTSLAGSYY358 pKa = 11.07ANWIILKK365 pKa = 9.69WIFLKK370 pKa = 10.99LGLEE374 pKa = 4.17VKK376 pKa = 9.86IWTFGDD382 pKa = 3.6DD383 pKa = 3.6SVVAIYY389 pKa = 10.07GQVDD393 pKa = 3.23TGGLMSQVVRR403 pKa = 11.84LAWEE407 pKa = 4.42GFGMNVSRR415 pKa = 11.84EE416 pKa = 3.9KK417 pKa = 11.02SYY419 pKa = 8.88VTCHH423 pKa = 6.28LVDD426 pKa = 3.49IADD429 pKa = 4.14DD430 pKa = 4.28PEE432 pKa = 4.18PQKK435 pKa = 11.16AGSFLSMYY443 pKa = 10.46FLQTPMGVRR452 pKa = 11.84PTRR455 pKa = 11.84PLQDD459 pKa = 3.17MYY461 pKa = 11.63EE462 pKa = 4.17LLLKK466 pKa = 10.31PEE468 pKa = 4.17KK469 pKa = 10.58CRR471 pKa = 11.84GTVEE475 pKa = 3.95WEE477 pKa = 4.18VVRR480 pKa = 11.84TSMAYY485 pKa = 10.06LVFYY489 pKa = 10.85YY490 pKa = 10.74NEE492 pKa = 3.69TARR495 pKa = 11.84YY496 pKa = 8.61VLEE499 pKa = 4.81EE500 pKa = 3.98YY501 pKa = 9.48WVWLHH506 pKa = 5.84RR507 pKa = 11.84VHH509 pKa = 7.84RR510 pKa = 11.84IPEE513 pKa = 4.15LTGTADD519 pKa = 4.26DD520 pKa = 3.87MALLRR525 pKa = 11.84EE526 pKa = 4.09MDD528 pKa = 3.9IPWSSFRR535 pKa = 11.84MEE537 pKa = 4.1WLNRR541 pKa = 11.84LAFHH545 pKa = 7.15GEE547 pKa = 3.82VEE549 pKa = 4.15LMYY552 pKa = 10.56KK553 pKa = 10.23YY554 pKa = 10.91GHH556 pKa = 6.11TKK558 pKa = 10.09FFPPVLWNAWYY569 pKa = 9.16QQYY572 pKa = 11.13DD573 pKa = 3.5EE574 pKa = 5.32SICGNEE580 pKa = 3.76IVPDD584 pKa = 3.54QRR586 pKa = 11.84APNDD590 pKa = 3.32
MM1 pKa = 6.68MVDD4 pKa = 4.2WIAEE8 pKa = 4.13NPSTVSEE15 pKa = 4.04ASGNGVKK22 pKa = 9.24EE23 pKa = 3.44WRR25 pKa = 11.84FFLKK29 pKa = 9.91HH30 pKa = 6.23CSWQRR35 pKa = 11.84RR36 pKa = 11.84LPGVSSLGHH45 pKa = 5.73SPAVRR50 pKa = 11.84YY51 pKa = 9.85RR52 pKa = 11.84YY53 pKa = 8.14VQPEE57 pKa = 4.15TSPSFLFSKK66 pKa = 10.54FLDD69 pKa = 3.89EE70 pKa = 4.55SQSSLGGIEE79 pKa = 4.24GPARR83 pKa = 11.84DD84 pKa = 3.65VLFSEE89 pKa = 5.33FVPPSEE95 pKa = 5.5DD96 pKa = 4.26DD97 pKa = 3.81LFEE100 pKa = 5.86HH101 pKa = 6.89IAGFGEE107 pKa = 4.29PGPDD111 pKa = 3.6GSHH114 pKa = 7.08LPGSDD119 pKa = 3.16FAQLFRR125 pKa = 11.84EE126 pKa = 4.41LRR128 pKa = 11.84RR129 pKa = 11.84LEE131 pKa = 3.97QWDD134 pKa = 3.97VPISGLLDD142 pKa = 3.07ATMLPKK148 pKa = 10.01IRR150 pKa = 11.84VPSATSPGIRR160 pKa = 11.84WKK162 pKa = 10.87KK163 pKa = 10.21LGYY166 pKa = 7.66KK167 pKa = 8.29TKK169 pKa = 10.53RR170 pKa = 11.84QALVPAVVEE179 pKa = 4.07ATRR182 pKa = 11.84ALNRR186 pKa = 11.84MVEE189 pKa = 4.19EE190 pKa = 4.21EE191 pKa = 4.1VAYY194 pKa = 8.04TVPPCGVAGRR204 pKa = 11.84GKK206 pKa = 10.09RR207 pKa = 11.84VNILEE212 pKa = 4.3GGSRR216 pKa = 11.84KK217 pKa = 9.63GKK219 pKa = 9.89KK220 pKa = 9.68DD221 pKa = 2.82GRR223 pKa = 11.84LIVMPDD229 pKa = 3.6LVRR232 pKa = 11.84HH233 pKa = 6.01LLGSLGSAPYY243 pKa = 9.48MSRR246 pKa = 11.84LRR248 pKa = 11.84GMDD251 pKa = 3.19HH252 pKa = 6.84SGGGVLLGMGPFQDD266 pKa = 3.9QYY268 pKa = 11.86QLIADD273 pKa = 4.41WATGASAFCFIDD285 pKa = 4.23FKK287 pKa = 11.43KK288 pKa = 10.49FDD290 pKa = 3.4QRR292 pKa = 11.84VPRR295 pKa = 11.84RR296 pKa = 11.84LLRR299 pKa = 11.84GVMNHH304 pKa = 5.77IRR306 pKa = 11.84SAFIGEE312 pKa = 4.25PGSRR316 pKa = 11.84AYY318 pKa = 9.44WEE320 pKa = 4.21SEE322 pKa = 3.48FRR324 pKa = 11.84HH325 pKa = 6.12LVDD328 pKa = 4.52TEE330 pKa = 3.66IAMPDD335 pKa = 3.75GSCYY339 pKa = 10.27RR340 pKa = 11.84KK341 pKa = 9.89RR342 pKa = 11.84GCVASGDD349 pKa = 3.84PWTSLAGSYY358 pKa = 11.07ANWIILKK365 pKa = 9.69WIFLKK370 pKa = 10.99LGLEE374 pKa = 4.17VKK376 pKa = 9.86IWTFGDD382 pKa = 3.6DD383 pKa = 3.6SVVAIYY389 pKa = 10.07GQVDD393 pKa = 3.23TGGLMSQVVRR403 pKa = 11.84LAWEE407 pKa = 4.42GFGMNVSRR415 pKa = 11.84EE416 pKa = 3.9KK417 pKa = 11.02SYY419 pKa = 8.88VTCHH423 pKa = 6.28LVDD426 pKa = 3.49IADD429 pKa = 4.14DD430 pKa = 4.28PEE432 pKa = 4.18PQKK435 pKa = 11.16AGSFLSMYY443 pKa = 10.46FLQTPMGVRR452 pKa = 11.84PTRR455 pKa = 11.84PLQDD459 pKa = 3.17MYY461 pKa = 11.63EE462 pKa = 4.17LLLKK466 pKa = 10.31PEE468 pKa = 4.17KK469 pKa = 10.58CRR471 pKa = 11.84GTVEE475 pKa = 3.95WEE477 pKa = 4.18VVRR480 pKa = 11.84TSMAYY485 pKa = 10.06LVFYY489 pKa = 10.85YY490 pKa = 10.74NEE492 pKa = 3.69TARR495 pKa = 11.84YY496 pKa = 8.61VLEE499 pKa = 4.81EE500 pKa = 3.98YY501 pKa = 9.48WVWLHH506 pKa = 5.84RR507 pKa = 11.84VHH509 pKa = 7.84RR510 pKa = 11.84IPEE513 pKa = 4.15LTGTADD519 pKa = 4.26DD520 pKa = 3.87MALLRR525 pKa = 11.84EE526 pKa = 4.09MDD528 pKa = 3.9IPWSSFRR535 pKa = 11.84MEE537 pKa = 4.1WLNRR541 pKa = 11.84LAFHH545 pKa = 7.15GEE547 pKa = 3.82VEE549 pKa = 4.15LMYY552 pKa = 10.56KK553 pKa = 10.23YY554 pKa = 10.91GHH556 pKa = 6.11TKK558 pKa = 10.09FFPPVLWNAWYY569 pKa = 9.16QQYY572 pKa = 11.13DD573 pKa = 3.5EE574 pKa = 5.32SICGNEE580 pKa = 3.76IVPDD584 pKa = 3.54QRR586 pKa = 11.84APNDD590 pKa = 3.32
Molecular weight: 66.96 kDa
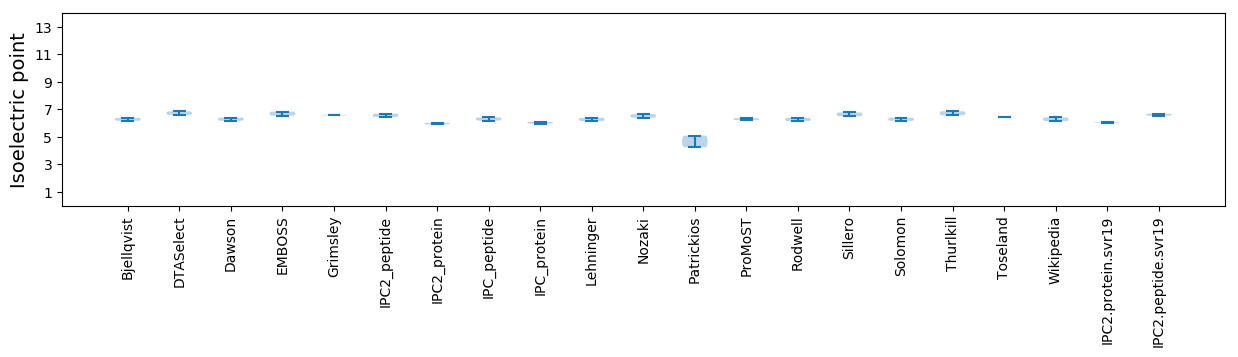
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0G4DJ74|A0A0G4DJ74_9VIRU Uncharacterized protein OS=Beauveria bassiana RNA virus 1 OX=1054649 PE=4 SV=1
MM1 pKa = 6.34MTPVARR7 pKa = 11.84NQEE10 pKa = 4.13RR11 pKa = 11.84ADD13 pKa = 3.49VSSVKK18 pKa = 10.12EE19 pKa = 4.18ATQSAPGTATNTGATEE35 pKa = 4.29RR36 pKa = 11.84PTSPAARR43 pKa = 11.84RR44 pKa = 11.84SSTPVRR50 pKa = 11.84PQVPPDD56 pKa = 3.83LEE58 pKa = 6.3AGLTLNMVTTRR69 pKa = 11.84TVPSHH74 pKa = 6.78PEE76 pKa = 3.17AVLRR80 pKa = 11.84VLQAFLGARR89 pKa = 11.84GVTTLPSSVRR99 pKa = 11.84DD100 pKa = 3.74LYY102 pKa = 10.48PSNWRR107 pKa = 11.84IGNTTIPVRR116 pKa = 11.84GSPEE120 pKa = 3.68SATRR124 pKa = 11.84LGEE127 pKa = 4.14VLPHH131 pKa = 6.19LRR133 pKa = 11.84GFNAKK138 pKa = 10.09GEE140 pKa = 4.2EE141 pKa = 4.09VHH143 pKa = 6.77WISEE147 pKa = 4.09GRR149 pKa = 11.84DD150 pKa = 3.2ASSVVRR156 pKa = 11.84LMAEE160 pKa = 3.38WDD162 pKa = 3.92SPVEE166 pKa = 3.56IAAAILADD174 pKa = 3.9VKK176 pKa = 11.21DD177 pKa = 3.91VDD179 pKa = 3.91VSKK182 pKa = 11.49DD183 pKa = 3.33DD184 pKa = 3.79VLAVAQRR191 pKa = 11.84SPQALEE197 pKa = 3.72QLKK200 pKa = 10.91KK201 pKa = 10.29SGKK204 pKa = 7.23EE205 pKa = 3.86FKK207 pKa = 10.47EE208 pKa = 4.34HH209 pKa = 4.16VQRR212 pKa = 11.84WLDD215 pKa = 3.6PVALRR220 pKa = 11.84RR221 pKa = 11.84SPEE224 pKa = 4.05FIAVSLRR231 pKa = 11.84YY232 pKa = 9.12KK233 pKa = 10.41AQLRR237 pKa = 11.84SLIDD241 pKa = 3.62EE242 pKa = 4.05QQKK245 pKa = 11.11AIVAARR251 pKa = 11.84KK252 pKa = 8.05ATAAVNRR259 pKa = 11.84ALGEE263 pKa = 3.87RR264 pKa = 11.84DD265 pKa = 3.08EE266 pKa = 4.68ALARR270 pKa = 11.84LDD272 pKa = 3.4PGYY275 pKa = 10.67VPKK278 pKa = 10.74RR279 pKa = 11.84ATAARR284 pKa = 11.84ALAEE288 pKa = 4.29FGIDD292 pKa = 3.68LDD294 pKa = 4.95GDD296 pKa = 4.04TEE298 pKa = 4.12MDD300 pKa = 3.99DD301 pKa = 3.83AAGAASDD308 pKa = 4.21ALRR311 pKa = 11.84DD312 pKa = 3.52LDD314 pKa = 4.03FF315 pKa = 5.87
MM1 pKa = 6.34MTPVARR7 pKa = 11.84NQEE10 pKa = 4.13RR11 pKa = 11.84ADD13 pKa = 3.49VSSVKK18 pKa = 10.12EE19 pKa = 4.18ATQSAPGTATNTGATEE35 pKa = 4.29RR36 pKa = 11.84PTSPAARR43 pKa = 11.84RR44 pKa = 11.84SSTPVRR50 pKa = 11.84PQVPPDD56 pKa = 3.83LEE58 pKa = 6.3AGLTLNMVTTRR69 pKa = 11.84TVPSHH74 pKa = 6.78PEE76 pKa = 3.17AVLRR80 pKa = 11.84VLQAFLGARR89 pKa = 11.84GVTTLPSSVRR99 pKa = 11.84DD100 pKa = 3.74LYY102 pKa = 10.48PSNWRR107 pKa = 11.84IGNTTIPVRR116 pKa = 11.84GSPEE120 pKa = 3.68SATRR124 pKa = 11.84LGEE127 pKa = 4.14VLPHH131 pKa = 6.19LRR133 pKa = 11.84GFNAKK138 pKa = 10.09GEE140 pKa = 4.2EE141 pKa = 4.09VHH143 pKa = 6.77WISEE147 pKa = 4.09GRR149 pKa = 11.84DD150 pKa = 3.2ASSVVRR156 pKa = 11.84LMAEE160 pKa = 3.38WDD162 pKa = 3.92SPVEE166 pKa = 3.56IAAAILADD174 pKa = 3.9VKK176 pKa = 11.21DD177 pKa = 3.91VDD179 pKa = 3.91VSKK182 pKa = 11.49DD183 pKa = 3.33DD184 pKa = 3.79VLAVAQRR191 pKa = 11.84SPQALEE197 pKa = 3.72QLKK200 pKa = 10.91KK201 pKa = 10.29SGKK204 pKa = 7.23EE205 pKa = 3.86FKK207 pKa = 10.47EE208 pKa = 4.34HH209 pKa = 4.16VQRR212 pKa = 11.84WLDD215 pKa = 3.6PVALRR220 pKa = 11.84RR221 pKa = 11.84SPEE224 pKa = 4.05FIAVSLRR231 pKa = 11.84YY232 pKa = 9.12KK233 pKa = 10.41AQLRR237 pKa = 11.84SLIDD241 pKa = 3.62EE242 pKa = 4.05QQKK245 pKa = 11.11AIVAARR251 pKa = 11.84KK252 pKa = 8.05ATAAVNRR259 pKa = 11.84ALGEE263 pKa = 3.87RR264 pKa = 11.84DD265 pKa = 3.08EE266 pKa = 4.68ALARR270 pKa = 11.84LDD272 pKa = 3.4PGYY275 pKa = 10.67VPKK278 pKa = 10.74RR279 pKa = 11.84ATAARR284 pKa = 11.84ALAEE288 pKa = 4.29FGIDD292 pKa = 3.68LDD294 pKa = 4.95GDD296 pKa = 4.04TEE298 pKa = 4.12MDD300 pKa = 3.99DD301 pKa = 3.83AAGAASDD308 pKa = 4.21ALRR311 pKa = 11.84DD312 pKa = 3.52LDD314 pKa = 4.03FF315 pKa = 5.87
Molecular weight: 34.05 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
905 |
315 |
590 |
452.5 |
50.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.95 ± 2.987 | 0.884 ± 0.495 |
5.967 ± 0.57 | 6.63 ± 0.021 |
3.757 ± 1.037 | 7.624 ± 1.247 |
1.878 ± 0.341 | 3.536 ± 0.38 |
3.978 ± 0.094 | 9.171 ± 0.158 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.762 ± 0.658 | 2.099 ± 0.069 |
6.409 ± 0.144 | 3.204 ± 0.161 |
7.956 ± 0.7 | 7.293 ± 0.005 |
4.53 ± 1.018 | 8.066 ± 0.461 |
2.541 ± 0.712 | 2.762 ± 1.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
