
Capybara microvirus Cap1_SP_263
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.13
Get precalculated fractions of proteins
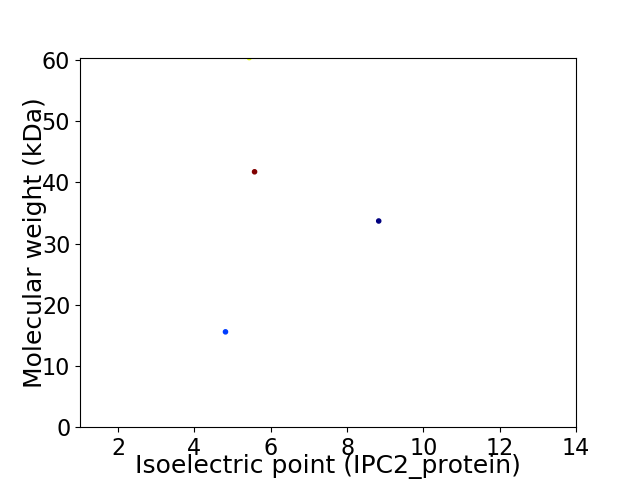
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
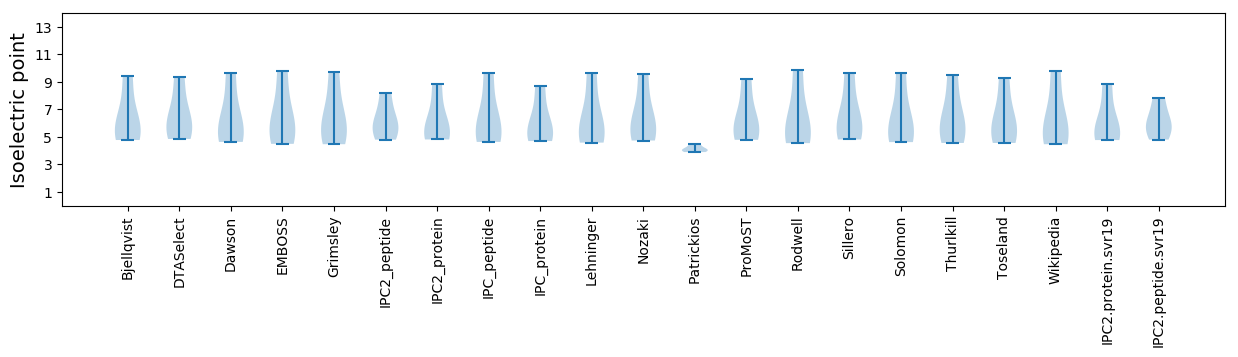
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W5I9|A0A4P8W5I9_9VIRU Replication initiator protein OS=Capybara microvirus Cap1_SP_263 OX=2585420 PE=4 SV=1
MM1 pKa = 7.65LNFLNFYY8 pKa = 10.72VKK10 pKa = 10.22FFFIMKK16 pKa = 9.64SISKK20 pKa = 9.06FHH22 pKa = 5.97YY23 pKa = 8.51NCKK26 pKa = 7.88KK27 pKa = 10.16QKK29 pKa = 10.27YY30 pKa = 7.9EE31 pKa = 4.12VNSGEE36 pKa = 4.31IKK38 pKa = 10.65VEE40 pKa = 3.9PCSCLSIKK48 pKa = 10.64DD49 pKa = 3.0IFEE52 pKa = 4.2RR53 pKa = 11.84FRR55 pKa = 11.84LGLPLDD61 pKa = 3.78SSVCINDD68 pKa = 3.68SYY70 pKa = 11.96DD71 pKa = 3.69EE72 pKa = 4.25EE73 pKa = 5.47SSTFLDD79 pKa = 3.3INTIKK84 pKa = 10.81QPDD87 pKa = 4.21LVSKK91 pKa = 10.42IDD93 pKa = 3.81LVASLHH99 pKa = 5.92EE100 pKa = 4.63FNKK103 pKa = 10.42SASEE107 pKa = 4.14TSVSRR112 pKa = 11.84SEE114 pKa = 3.97MGVEE118 pKa = 4.16EE119 pKa = 4.1VQPLNEE125 pKa = 4.19SAEE128 pKa = 4.03EE129 pKa = 3.82SDD131 pKa = 5.07NIEE134 pKa = 4.05KK135 pKa = 10.77SS136 pKa = 3.26
MM1 pKa = 7.65LNFLNFYY8 pKa = 10.72VKK10 pKa = 10.22FFFIMKK16 pKa = 9.64SISKK20 pKa = 9.06FHH22 pKa = 5.97YY23 pKa = 8.51NCKK26 pKa = 7.88KK27 pKa = 10.16QKK29 pKa = 10.27YY30 pKa = 7.9EE31 pKa = 4.12VNSGEE36 pKa = 4.31IKK38 pKa = 10.65VEE40 pKa = 3.9PCSCLSIKK48 pKa = 10.64DD49 pKa = 3.0IFEE52 pKa = 4.2RR53 pKa = 11.84FRR55 pKa = 11.84LGLPLDD61 pKa = 3.78SSVCINDD68 pKa = 3.68SYY70 pKa = 11.96DD71 pKa = 3.69EE72 pKa = 4.25EE73 pKa = 5.47SSTFLDD79 pKa = 3.3INTIKK84 pKa = 10.81QPDD87 pKa = 4.21LVSKK91 pKa = 10.42IDD93 pKa = 3.81LVASLHH99 pKa = 5.92EE100 pKa = 4.63FNKK103 pKa = 10.42SASEE107 pKa = 4.14TSVSRR112 pKa = 11.84SEE114 pKa = 3.97MGVEE118 pKa = 4.16EE119 pKa = 4.1VQPLNEE125 pKa = 4.19SAEE128 pKa = 4.03EE129 pKa = 3.82SDD131 pKa = 5.07NIEE134 pKa = 4.05KK135 pKa = 10.77SS136 pKa = 3.26
Molecular weight: 15.58 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W5R5|A0A4P8W5R5_9VIRU Major capsid protein OS=Capybara microvirus Cap1_SP_263 OX=2585420 PE=3 SV=1
MM1 pKa = 7.68CLFPKK6 pKa = 9.85QVRR9 pKa = 11.84VTLKK13 pKa = 10.43DD14 pKa = 3.35GHH16 pKa = 5.92SVPMSVPCGKK26 pKa = 10.67CIDD29 pKa = 4.01CLQLRR34 pKa = 11.84THH36 pKa = 6.43DD37 pKa = 3.04WEE39 pKa = 4.76LRR41 pKa = 11.84MKK43 pKa = 11.09YY44 pKa = 10.07EE45 pKa = 4.6LLNSNYY51 pKa = 10.54GLFVTLTYY59 pKa = 11.25DD60 pKa = 4.44DD61 pKa = 5.63FSCPSLGVEE70 pKa = 4.15VSHH73 pKa = 5.4VQKK76 pKa = 10.72FIKK79 pKa = 10.07RR80 pKa = 11.84LRR82 pKa = 11.84KK83 pKa = 8.97HH84 pKa = 5.05GHH86 pKa = 4.56IFKK89 pKa = 9.97YY90 pKa = 10.46YY91 pKa = 10.49CIGEE95 pKa = 4.37YY96 pKa = 9.42GTSTFRR102 pKa = 11.84PHH104 pKa = 4.74YY105 pKa = 9.77HH106 pKa = 7.12LIFFVTSDD114 pKa = 3.12SHH116 pKa = 8.29SINLSLDD123 pKa = 3.38INKK126 pKa = 9.44SWPYY130 pKa = 11.03GFTQVKK136 pKa = 8.43PCSFGRR142 pKa = 11.84IHH144 pKa = 6.46YY145 pKa = 9.47CIAYY149 pKa = 10.33LIGQKK154 pKa = 10.39SPLGKK159 pKa = 10.1NKK161 pKa = 9.78PFTIMSKK168 pKa = 10.07RR169 pKa = 11.84PSIGLCALDD178 pKa = 4.7DD179 pKa = 3.87KK180 pKa = 11.46TFVSQLKK187 pKa = 8.67STNFEE192 pKa = 3.96YY193 pKa = 10.61TFRR196 pKa = 11.84NGHH199 pKa = 5.73KK200 pKa = 9.54VHH202 pKa = 5.84VPRR205 pKa = 11.84YY206 pKa = 8.39YY207 pKa = 10.19RR208 pKa = 11.84RR209 pKa = 11.84KK210 pKa = 10.57LNDD213 pKa = 3.34KK214 pKa = 10.69DD215 pKa = 3.48FTLIDD220 pKa = 4.67FYY222 pKa = 11.56LLKK225 pKa = 10.44CKK227 pKa = 10.16HH228 pKa = 6.37HH229 pKa = 5.78EE230 pKa = 4.28QFLKK234 pKa = 10.53DD235 pKa = 3.5YY236 pKa = 10.97KK237 pKa = 11.03SYY239 pKa = 11.4LNSKK243 pKa = 9.75HH244 pKa = 6.8LDD246 pKa = 3.55FSKK249 pKa = 10.49HH250 pKa = 4.84YY251 pKa = 11.15NNLDD255 pKa = 3.84LIKK258 pKa = 10.34DD259 pKa = 4.02FKK261 pKa = 10.71ISQSIYY267 pKa = 10.44NFSHH271 pKa = 7.83DD272 pKa = 4.57EE273 pKa = 3.73ILKK276 pKa = 10.06NRR278 pKa = 11.84NKK280 pKa = 10.38KK281 pKa = 9.94LKK283 pKa = 10.46NDD285 pKa = 3.41
MM1 pKa = 7.68CLFPKK6 pKa = 9.85QVRR9 pKa = 11.84VTLKK13 pKa = 10.43DD14 pKa = 3.35GHH16 pKa = 5.92SVPMSVPCGKK26 pKa = 10.67CIDD29 pKa = 4.01CLQLRR34 pKa = 11.84THH36 pKa = 6.43DD37 pKa = 3.04WEE39 pKa = 4.76LRR41 pKa = 11.84MKK43 pKa = 11.09YY44 pKa = 10.07EE45 pKa = 4.6LLNSNYY51 pKa = 10.54GLFVTLTYY59 pKa = 11.25DD60 pKa = 4.44DD61 pKa = 5.63FSCPSLGVEE70 pKa = 4.15VSHH73 pKa = 5.4VQKK76 pKa = 10.72FIKK79 pKa = 10.07RR80 pKa = 11.84LRR82 pKa = 11.84KK83 pKa = 8.97HH84 pKa = 5.05GHH86 pKa = 4.56IFKK89 pKa = 9.97YY90 pKa = 10.46YY91 pKa = 10.49CIGEE95 pKa = 4.37YY96 pKa = 9.42GTSTFRR102 pKa = 11.84PHH104 pKa = 4.74YY105 pKa = 9.77HH106 pKa = 7.12LIFFVTSDD114 pKa = 3.12SHH116 pKa = 8.29SINLSLDD123 pKa = 3.38INKK126 pKa = 9.44SWPYY130 pKa = 11.03GFTQVKK136 pKa = 8.43PCSFGRR142 pKa = 11.84IHH144 pKa = 6.46YY145 pKa = 9.47CIAYY149 pKa = 10.33LIGQKK154 pKa = 10.39SPLGKK159 pKa = 10.1NKK161 pKa = 9.78PFTIMSKK168 pKa = 10.07RR169 pKa = 11.84PSIGLCALDD178 pKa = 4.7DD179 pKa = 3.87KK180 pKa = 11.46TFVSQLKK187 pKa = 8.67STNFEE192 pKa = 3.96YY193 pKa = 10.61TFRR196 pKa = 11.84NGHH199 pKa = 5.73KK200 pKa = 9.54VHH202 pKa = 5.84VPRR205 pKa = 11.84YY206 pKa = 8.39YY207 pKa = 10.19RR208 pKa = 11.84RR209 pKa = 11.84KK210 pKa = 10.57LNDD213 pKa = 3.34KK214 pKa = 10.69DD215 pKa = 3.48FTLIDD220 pKa = 4.67FYY222 pKa = 11.56LLKK225 pKa = 10.44CKK227 pKa = 10.16HH228 pKa = 6.37HH229 pKa = 5.78EE230 pKa = 4.28QFLKK234 pKa = 10.53DD235 pKa = 3.5YY236 pKa = 10.97KK237 pKa = 11.03SYY239 pKa = 11.4LNSKK243 pKa = 9.75HH244 pKa = 6.8LDD246 pKa = 3.55FSKK249 pKa = 10.49HH250 pKa = 4.84YY251 pKa = 11.15NNLDD255 pKa = 3.84LIKK258 pKa = 10.34DD259 pKa = 4.02FKK261 pKa = 10.71ISQSIYY267 pKa = 10.44NFSHH271 pKa = 7.83DD272 pKa = 4.57EE273 pKa = 3.73ILKK276 pKa = 10.06NRR278 pKa = 11.84NKK280 pKa = 10.38KK281 pKa = 9.94LKK283 pKa = 10.46NDD285 pKa = 3.41
Molecular weight: 33.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1315 |
136 |
531 |
328.8 |
37.86 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.411 ± 1.508 | 1.369 ± 0.806 |
5.779 ± 0.637 | 6.008 ± 1.365 |
6.236 ± 1.05 | 5.551 ± 1.26 |
2.586 ± 0.91 | 5.399 ± 0.474 |
6.996 ± 1.589 | 10.038 ± 0.901 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.293 ± 0.196 | 7.605 ± 1.151 |
3.878 ± 0.599 | 5.095 ± 1.323 |
4.487 ± 0.881 | 8.061 ± 1.399 |
4.487 ± 0.457 | 4.867 ± 0.698 |
0.913 ± 0.226 | 4.943 ± 0.536 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
