
JC polyomavirus (JCPyV) (JCV)
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; Betapolyomavirus; Human polyomavirus 2
Average proteome isoelectric point is 6.99
Get precalculated fractions of proteins
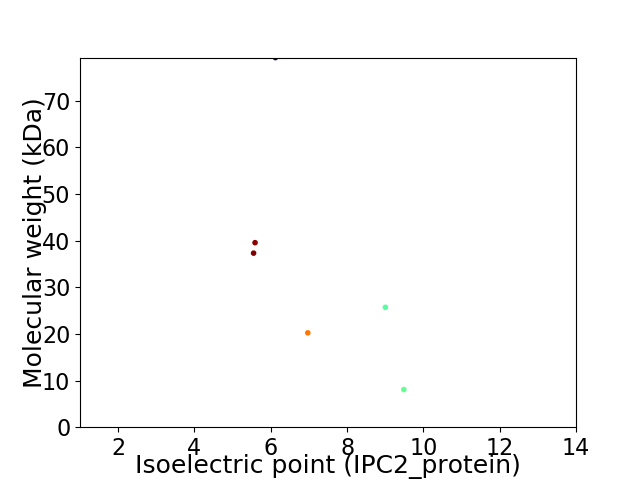
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
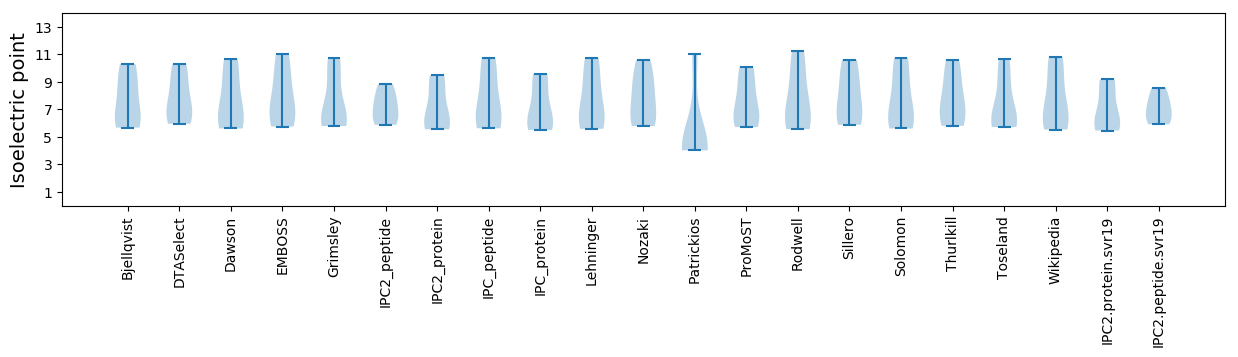
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|O92720|O92720_POVJC Minor capsid protein OS=JC polyomavirus OX=10632 GN=VP3 PE=3 SV=1
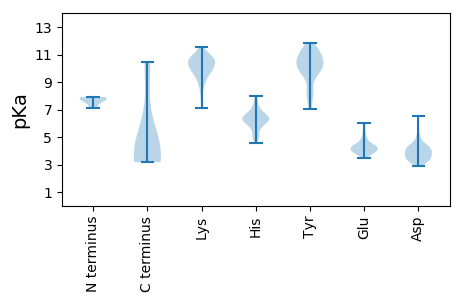
MM1 pKa = 7.14GAALALLGDD10 pKa = 4.3LVATVSEE17 pKa = 4.25AAAATGFSVAEE28 pKa = 4.01IAAGEE33 pKa = 3.91AAATIEE39 pKa = 4.3VEE41 pKa = 4.28IASLATVEE49 pKa = 5.49GITSTSEE56 pKa = 3.82AIAAIGLTPEE66 pKa = 4.45TYY68 pKa = 10.83AVITGAPGAVAGFAALVQTVTGGSAIAQLGYY99 pKa = 10.77RR100 pKa = 11.84FFADD104 pKa = 3.05WDD106 pKa = 4.17HH107 pKa = 6.5KK108 pKa = 11.14VSTVGLFQQPAMALQLFNPEE128 pKa = 4.35DD129 pKa = 4.17YY130 pKa = 11.39YY131 pKa = 11.45DD132 pKa = 3.52ILFPGVNAFVNNIHH146 pKa = 6.25YY147 pKa = 10.37LDD149 pKa = 4.1PRR151 pKa = 11.84HH152 pKa = 6.33WGPSLFSTISQAFWNLVRR170 pKa = 11.84DD171 pKa = 4.39DD172 pKa = 5.17LPALTSQEE180 pKa = 3.86IQRR183 pKa = 11.84RR184 pKa = 11.84TQKK187 pKa = 11.02LFVEE191 pKa = 4.21NLARR195 pKa = 11.84FLEE198 pKa = 4.37EE199 pKa = 3.93TTWAIVNSPANLYY212 pKa = 10.82NYY214 pKa = 9.9ISDD217 pKa = 4.06YY218 pKa = 10.43YY219 pKa = 11.38SRR221 pKa = 11.84LSPVRR226 pKa = 11.84PSMVRR231 pKa = 11.84QVAQRR236 pKa = 11.84EE237 pKa = 4.28GTYY240 pKa = 10.19ISFGHH245 pKa = 6.57SYY247 pKa = 8.19TQSIDD252 pKa = 3.57DD253 pKa = 4.19ADD255 pKa = 4.52SIQEE259 pKa = 3.85VTQRR263 pKa = 11.84LDD265 pKa = 3.3LKK267 pKa = 9.91TPNVQSGEE275 pKa = 4.07FIEE278 pKa = 5.69RR279 pKa = 11.84SIAPGGANQRR289 pKa = 11.84SAPQWMLPLLLGLYY303 pKa = 8.21GTVTPALAAYY313 pKa = 9.61EE314 pKa = 4.43DD315 pKa = 4.33GPNKK319 pKa = 10.02KK320 pKa = 9.5KK321 pKa = 10.16RR322 pKa = 11.84RR323 pKa = 11.84KK324 pKa = 8.4EE325 pKa = 3.86GPRR328 pKa = 11.84ASSKK332 pKa = 9.28TSYY335 pKa = 10.17KK336 pKa = 9.95RR337 pKa = 11.84RR338 pKa = 11.84SRR340 pKa = 11.84SSRR343 pKa = 11.84SS344 pKa = 3.18
MM1 pKa = 7.14GAALALLGDD10 pKa = 4.3LVATVSEE17 pKa = 4.25AAAATGFSVAEE28 pKa = 4.01IAAGEE33 pKa = 3.91AAATIEE39 pKa = 4.3VEE41 pKa = 4.28IASLATVEE49 pKa = 5.49GITSTSEE56 pKa = 3.82AIAAIGLTPEE66 pKa = 4.45TYY68 pKa = 10.83AVITGAPGAVAGFAALVQTVTGGSAIAQLGYY99 pKa = 10.77RR100 pKa = 11.84FFADD104 pKa = 3.05WDD106 pKa = 4.17HH107 pKa = 6.5KK108 pKa = 11.14VSTVGLFQQPAMALQLFNPEE128 pKa = 4.35DD129 pKa = 4.17YY130 pKa = 11.39YY131 pKa = 11.45DD132 pKa = 3.52ILFPGVNAFVNNIHH146 pKa = 6.25YY147 pKa = 10.37LDD149 pKa = 4.1PRR151 pKa = 11.84HH152 pKa = 6.33WGPSLFSTISQAFWNLVRR170 pKa = 11.84DD171 pKa = 4.39DD172 pKa = 5.17LPALTSQEE180 pKa = 3.86IQRR183 pKa = 11.84RR184 pKa = 11.84TQKK187 pKa = 11.02LFVEE191 pKa = 4.21NLARR195 pKa = 11.84FLEE198 pKa = 4.37EE199 pKa = 3.93TTWAIVNSPANLYY212 pKa = 10.82NYY214 pKa = 9.9ISDD217 pKa = 4.06YY218 pKa = 10.43YY219 pKa = 11.38SRR221 pKa = 11.84LSPVRR226 pKa = 11.84PSMVRR231 pKa = 11.84QVAQRR236 pKa = 11.84EE237 pKa = 4.28GTYY240 pKa = 10.19ISFGHH245 pKa = 6.57SYY247 pKa = 8.19TQSIDD252 pKa = 3.57DD253 pKa = 4.19ADD255 pKa = 4.52SIQEE259 pKa = 3.85VTQRR263 pKa = 11.84LDD265 pKa = 3.3LKK267 pKa = 9.91TPNVQSGEE275 pKa = 4.07FIEE278 pKa = 5.69RR279 pKa = 11.84SIAPGGANQRR289 pKa = 11.84SAPQWMLPLLLGLYY303 pKa = 8.21GTVTPALAAYY313 pKa = 9.61EE314 pKa = 4.43DD315 pKa = 4.33GPNKK319 pKa = 10.02KK320 pKa = 9.5KK321 pKa = 10.16RR322 pKa = 11.84RR323 pKa = 11.84KK324 pKa = 8.4EE325 pKa = 3.86GPRR328 pKa = 11.84ASSKK332 pKa = 9.28TSYY335 pKa = 10.17KK336 pKa = 9.95RR337 pKa = 11.84RR338 pKa = 11.84SRR340 pKa = 11.84SSRR343 pKa = 11.84SS344 pKa = 3.18
Molecular weight: 37.34 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|O55864|O55864_POVJC Minor capsid protein OS=JC polyomavirus OX=10632 GN=VP2 PE=3 SV=1
MM1 pKa = 7.53VLRR4 pKa = 11.84QLSRR8 pKa = 11.84KK9 pKa = 9.98ASVKK13 pKa = 10.15VSKK16 pKa = 8.76TWSGTKK22 pKa = 9.78KK23 pKa = 9.32RR24 pKa = 11.84AQRR27 pKa = 11.84ILIFLLEE34 pKa = 4.11FLLDD38 pKa = 4.07FCTGEE43 pKa = 4.54DD44 pKa = 3.53SVDD47 pKa = 3.0GKK49 pKa = 10.74KK50 pKa = 9.81RR51 pKa = 11.84QRR53 pKa = 11.84HH54 pKa = 5.41SGLTQQTYY62 pKa = 10.71SALPEE67 pKa = 4.2PKK69 pKa = 9.57ATT71 pKa = 3.91
MM1 pKa = 7.53VLRR4 pKa = 11.84QLSRR8 pKa = 11.84KK9 pKa = 9.98ASVKK13 pKa = 10.15VSKK16 pKa = 8.76TWSGTKK22 pKa = 9.78KK23 pKa = 9.32RR24 pKa = 11.84AQRR27 pKa = 11.84ILIFLLEE34 pKa = 4.11FLLDD38 pKa = 4.07FCTGEE43 pKa = 4.54DD44 pKa = 3.53SVDD47 pKa = 3.0GKK49 pKa = 10.74KK50 pKa = 9.81RR51 pKa = 11.84QRR53 pKa = 11.84HH54 pKa = 5.41SGLTQQTYY62 pKa = 10.71SALPEE67 pKa = 4.2PKK69 pKa = 9.57ATT71 pKa = 3.91
Molecular weight: 8.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1854 |
71 |
688 |
309.0 |
35.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.365 ± 1.593 | 2.05 ± 0.757 |
5.609 ± 0.428 | 6.365 ± 0.554 |
4.639 ± 0.425 | 6.203 ± 0.468 |
2.211 ± 0.437 | 3.883 ± 0.494 |
6.419 ± 1.201 | 9.277 ± 0.34 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.805 ± 0.508 | 4.369 ± 0.354 |
5.34 ± 0.473 | 4.585 ± 0.347 |
5.609 ± 0.641 | 7.012 ± 0.737 |
5.717 ± 0.635 | 6.365 ± 0.514 |
1.618 ± 0.259 | 3.56 ± 0.345 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
