
Circovirus-like genome SAR-B
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 8.79
Get precalculated fractions of proteins
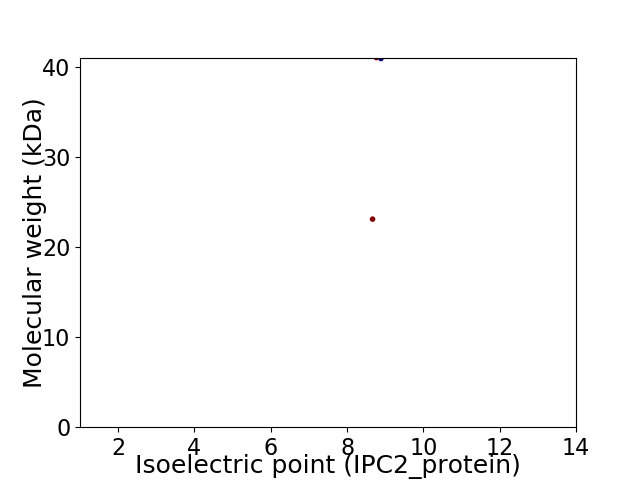
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C6GIJ3|C6GIJ3_9VIRU Putative Rep protein OS=Circovirus-like genome SAR-B OX=642259 PE=4 SV=1
MM1 pKa = 7.78ASQHH5 pKa = 5.09GWVFTLNNPSIEE17 pKa = 4.12EE18 pKa = 4.1YY19 pKa = 9.82PLKK22 pKa = 10.78FEE24 pKa = 4.33EE25 pKa = 4.6TSSGRR30 pKa = 11.84RR31 pKa = 11.84YY32 pKa = 9.98PIAKK36 pKa = 9.15WEE38 pKa = 4.15LPRR41 pKa = 11.84ATLAFLGCGRR51 pKa = 11.84EE52 pKa = 3.99RR53 pKa = 11.84GQSNTPHH60 pKa = 6.27LQGLLISSRR69 pKa = 11.84PVSLKK74 pKa = 10.31SLKK77 pKa = 10.23KK78 pKa = 10.32LNPRR82 pKa = 11.84AHH84 pKa = 6.73WEE86 pKa = 3.79PMRR89 pKa = 11.84GSFKK93 pKa = 10.22QARR96 pKa = 11.84EE97 pKa = 3.94YY98 pKa = 10.97CEE100 pKa = 4.7KK101 pKa = 10.57EE102 pKa = 4.05GKK104 pKa = 8.93FVQWTKK110 pKa = 10.64SGEE113 pKa = 4.46SVASLSRR120 pKa = 11.84YY121 pKa = 7.97VQTDD125 pKa = 3.3LEE127 pKa = 4.53IFQMIQQSDD136 pKa = 3.33TDD138 pKa = 4.29LNKK141 pKa = 10.71LSVTMLKK148 pKa = 9.75QEE150 pKa = 4.19KK151 pKa = 10.04KK152 pKa = 10.36LAEE155 pKa = 3.84LSKK158 pKa = 10.65RR159 pKa = 11.84IEE161 pKa = 4.79DD162 pKa = 3.6LTKK165 pKa = 10.62LQSHH169 pKa = 7.18FLLKK173 pKa = 10.05QDD175 pKa = 3.81NFQNSILKK183 pKa = 10.22LLSQKK188 pKa = 10.63KK189 pKa = 9.61ILNTLSDD196 pKa = 3.6EE197 pKa = 4.37DD198 pKa = 5.01LII200 pKa = 5.98
MM1 pKa = 7.78ASQHH5 pKa = 5.09GWVFTLNNPSIEE17 pKa = 4.12EE18 pKa = 4.1YY19 pKa = 9.82PLKK22 pKa = 10.78FEE24 pKa = 4.33EE25 pKa = 4.6TSSGRR30 pKa = 11.84RR31 pKa = 11.84YY32 pKa = 9.98PIAKK36 pKa = 9.15WEE38 pKa = 4.15LPRR41 pKa = 11.84ATLAFLGCGRR51 pKa = 11.84EE52 pKa = 3.99RR53 pKa = 11.84GQSNTPHH60 pKa = 6.27LQGLLISSRR69 pKa = 11.84PVSLKK74 pKa = 10.31SLKK77 pKa = 10.23KK78 pKa = 10.32LNPRR82 pKa = 11.84AHH84 pKa = 6.73WEE86 pKa = 3.79PMRR89 pKa = 11.84GSFKK93 pKa = 10.22QARR96 pKa = 11.84EE97 pKa = 3.94YY98 pKa = 10.97CEE100 pKa = 4.7KK101 pKa = 10.57EE102 pKa = 4.05GKK104 pKa = 8.93FVQWTKK110 pKa = 10.64SGEE113 pKa = 4.46SVASLSRR120 pKa = 11.84YY121 pKa = 7.97VQTDD125 pKa = 3.3LEE127 pKa = 4.53IFQMIQQSDD136 pKa = 3.33TDD138 pKa = 4.29LNKK141 pKa = 10.71LSVTMLKK148 pKa = 9.75QEE150 pKa = 4.19KK151 pKa = 10.04KK152 pKa = 10.36LAEE155 pKa = 3.84LSKK158 pKa = 10.65RR159 pKa = 11.84IEE161 pKa = 4.79DD162 pKa = 3.6LTKK165 pKa = 10.62LQSHH169 pKa = 7.18FLLKK173 pKa = 10.05QDD175 pKa = 3.81NFQNSILKK183 pKa = 10.22LLSQKK188 pKa = 10.63KK189 pKa = 9.61ILNTLSDD196 pKa = 3.6EE197 pKa = 4.37DD198 pKa = 5.01LII200 pKa = 5.98
Molecular weight: 23.08 kDa
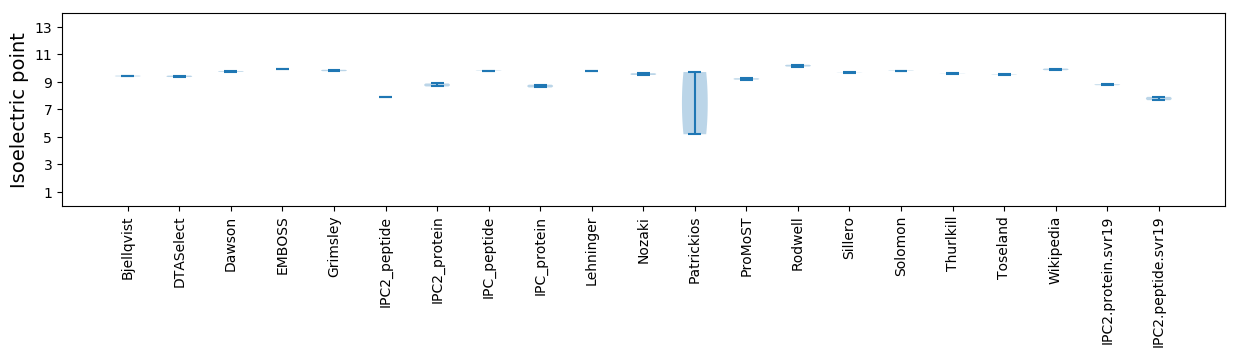
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C6GIJ3|C6GIJ3_9VIRU Putative Rep protein OS=Circovirus-like genome SAR-B OX=642259 PE=4 SV=1
MM1 pKa = 8.04VRR3 pKa = 11.84MTTGRR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 8.12TFRR13 pKa = 11.84SIKK16 pKa = 10.09KK17 pKa = 8.8VATRR21 pKa = 11.84GKK23 pKa = 10.36DD24 pKa = 3.3VIDD27 pKa = 3.74TVIEE31 pKa = 3.88ATEE34 pKa = 3.97KK35 pKa = 10.91GKK37 pKa = 10.78EE38 pKa = 3.95FVDD41 pKa = 5.14NISQMPVKK49 pKa = 10.37EE50 pKa = 4.3VMTTAAKK57 pKa = 10.14LAQSKK62 pKa = 10.04KK63 pKa = 10.29SPIGATGLNLLPKK76 pKa = 9.19EE77 pKa = 4.06ARR79 pKa = 11.84SVVSSTDD86 pKa = 3.42AIGDD90 pKa = 3.92FTSSASMFMYY100 pKa = 10.43RR101 pKa = 11.84PPRR104 pKa = 11.84KK105 pKa = 9.54SNRR108 pKa = 11.84AGFTKK113 pKa = 10.73YY114 pKa = 9.07QLKK117 pKa = 9.61TGATRR122 pKa = 11.84TNSSLSNQVGTSDD135 pKa = 4.2MNILDD140 pKa = 5.32AIQVEE145 pKa = 4.57NNPDD149 pKa = 3.26TDD151 pKa = 3.49AKK153 pKa = 10.86YY154 pKa = 11.21SNLSVKK160 pKa = 10.02EE161 pKa = 3.87AFDD164 pKa = 3.99NYY166 pKa = 10.83LLAAGLKK173 pKa = 8.53DD174 pKa = 3.71TAGTSYY180 pKa = 10.66DD181 pKa = 3.42QKK183 pKa = 10.58IQQTSIHH190 pKa = 5.99VSSLQSEE197 pKa = 4.17LVITNNNNDD206 pKa = 3.32TVMVDD211 pKa = 3.65LYY213 pKa = 11.18EE214 pKa = 4.93LVPQHH219 pKa = 6.09TLGPSQYY226 pKa = 10.54VNEE229 pKa = 4.27NQATGYY235 pKa = 8.37MSPVWTWTQGLSDD248 pKa = 3.73TVMVDD253 pKa = 3.41DD254 pKa = 5.04ALSRR258 pKa = 11.84SYY260 pKa = 10.8FQANPFNSSLFSRR273 pKa = 11.84TWKK276 pKa = 9.86IVKK279 pKa = 8.45QVRR282 pKa = 11.84ANISGGSTHH291 pKa = 5.75RR292 pKa = 11.84HH293 pKa = 4.87KK294 pKa = 10.68SAYY297 pKa = 8.92MINKK301 pKa = 6.71TVSYY305 pKa = 10.82PEE307 pKa = 3.85MAQFTTRR314 pKa = 11.84GGKK317 pKa = 8.71FAGWNPTFMIVQQGVPTSAAQEE339 pKa = 4.4GVASSITVRR348 pKa = 11.84MNAQLNYY355 pKa = 9.01EE356 pKa = 4.21ASAVEE361 pKa = 3.86QARR364 pKa = 11.84VIVFDD369 pKa = 4.05SKK371 pKa = 9.86TT372 pKa = 3.19
MM1 pKa = 8.04VRR3 pKa = 11.84MTTGRR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 8.12TFRR13 pKa = 11.84SIKK16 pKa = 10.09KK17 pKa = 8.8VATRR21 pKa = 11.84GKK23 pKa = 10.36DD24 pKa = 3.3VIDD27 pKa = 3.74TVIEE31 pKa = 3.88ATEE34 pKa = 3.97KK35 pKa = 10.91GKK37 pKa = 10.78EE38 pKa = 3.95FVDD41 pKa = 5.14NISQMPVKK49 pKa = 10.37EE50 pKa = 4.3VMTTAAKK57 pKa = 10.14LAQSKK62 pKa = 10.04KK63 pKa = 10.29SPIGATGLNLLPKK76 pKa = 9.19EE77 pKa = 4.06ARR79 pKa = 11.84SVVSSTDD86 pKa = 3.42AIGDD90 pKa = 3.92FTSSASMFMYY100 pKa = 10.43RR101 pKa = 11.84PPRR104 pKa = 11.84KK105 pKa = 9.54SNRR108 pKa = 11.84AGFTKK113 pKa = 10.73YY114 pKa = 9.07QLKK117 pKa = 9.61TGATRR122 pKa = 11.84TNSSLSNQVGTSDD135 pKa = 4.2MNILDD140 pKa = 5.32AIQVEE145 pKa = 4.57NNPDD149 pKa = 3.26TDD151 pKa = 3.49AKK153 pKa = 10.86YY154 pKa = 11.21SNLSVKK160 pKa = 10.02EE161 pKa = 3.87AFDD164 pKa = 3.99NYY166 pKa = 10.83LLAAGLKK173 pKa = 8.53DD174 pKa = 3.71TAGTSYY180 pKa = 10.66DD181 pKa = 3.42QKK183 pKa = 10.58IQQTSIHH190 pKa = 5.99VSSLQSEE197 pKa = 4.17LVITNNNNDD206 pKa = 3.32TVMVDD211 pKa = 3.65LYY213 pKa = 11.18EE214 pKa = 4.93LVPQHH219 pKa = 6.09TLGPSQYY226 pKa = 10.54VNEE229 pKa = 4.27NQATGYY235 pKa = 8.37MSPVWTWTQGLSDD248 pKa = 3.73TVMVDD253 pKa = 3.41DD254 pKa = 5.04ALSRR258 pKa = 11.84SYY260 pKa = 10.8FQANPFNSSLFSRR273 pKa = 11.84TWKK276 pKa = 9.86IVKK279 pKa = 8.45QVRR282 pKa = 11.84ANISGGSTHH291 pKa = 5.75RR292 pKa = 11.84HH293 pKa = 4.87KK294 pKa = 10.68SAYY297 pKa = 8.92MINKK301 pKa = 6.71TVSYY305 pKa = 10.82PEE307 pKa = 3.85MAQFTTRR314 pKa = 11.84GGKK317 pKa = 8.71FAGWNPTFMIVQQGVPTSAAQEE339 pKa = 4.4GVASSITVRR348 pKa = 11.84MNAQLNYY355 pKa = 9.01EE356 pKa = 4.21ASAVEE361 pKa = 3.86QARR364 pKa = 11.84VIVFDD369 pKa = 4.05SKK371 pKa = 9.86TT372 pKa = 3.19
Molecular weight: 40.91 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
572 |
200 |
372 |
286.0 |
31.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.818 ± 1.505 | 0.35 ± 0.347 |
4.371 ± 0.465 | 5.245 ± 1.471 |
3.671 ± 0.176 | 5.245 ± 0.398 |
1.399 ± 0.321 | 4.545 ± 0.024 |
7.517 ± 1.059 | 8.392 ± 2.995 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.147 ± 0.612 | 5.42 ± 0.758 |
3.671 ± 0.176 | 6.119 ± 0.471 |
4.895 ± 0.323 | 10.49 ± 0.006 |
8.042 ± 1.625 | 6.469 ± 1.852 |
1.399 ± 0.321 | 2.797 ± 0.426 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
