
Wuhan House Fly Virus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alpharhabdovirinae; Sigmavirus; Wuhan sigmavirus
Average proteome isoelectric point is 6.45
Get precalculated fractions of proteins
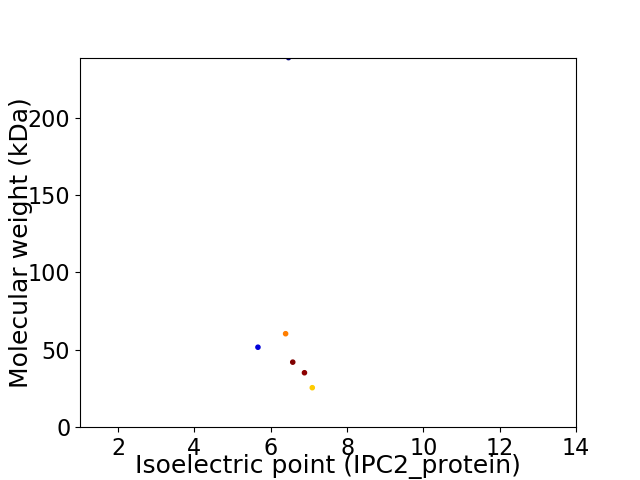
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B5KXL1|A0A0B5KXL1_9RHAB GDP polyribonucleotidyltransferase OS=Wuhan House Fly Virus 1 OX=1608104 GN=L PE=4 SV=1
MM1 pKa = 7.97DD2 pKa = 4.47VLNLTDD8 pKa = 3.42QEE10 pKa = 4.59TNEE13 pKa = 4.17FLRR16 pKa = 11.84VRR18 pKa = 11.84AIQIEE23 pKa = 4.34KK24 pKa = 8.57TGEE27 pKa = 3.78YY28 pKa = 9.61VYY30 pKa = 10.71PWFEE34 pKa = 3.84QHH36 pKa = 6.44NSKK39 pKa = 10.7PPIKK43 pKa = 9.92IPKK46 pKa = 9.27VDD48 pKa = 3.94DD49 pKa = 3.63AAGARR54 pKa = 11.84LRR56 pKa = 11.84AITKK60 pKa = 8.99GFASGVICSVQTASAFLSWKK80 pKa = 7.7MKK82 pKa = 10.4RR83 pKa = 11.84MTARR87 pKa = 11.84VDD89 pKa = 4.25SVWKK93 pKa = 10.45SYY95 pKa = 10.71NQLIGDD101 pKa = 4.58PNADD105 pKa = 3.24ITPLSLLSVEE115 pKa = 4.7EE116 pKa = 4.29VGTPTISGNEE126 pKa = 4.05VSSSEE131 pKa = 4.93ADD133 pKa = 3.52EE134 pKa = 4.16KK135 pKa = 10.97WIIGYY140 pKa = 8.51ILCLYY145 pKa = 10.56RR146 pKa = 11.84LGRR149 pKa = 11.84VNDD152 pKa = 3.49ANYY155 pKa = 10.53RR156 pKa = 11.84GDD158 pKa = 4.52LIKK161 pKa = 10.87SMKK164 pKa = 9.75PILEE168 pKa = 4.27AQGMPNTTNLLADD181 pKa = 4.24CGKK184 pKa = 7.14TTAWCQCPVFRR195 pKa = 11.84KK196 pKa = 9.86LVAAIDD202 pKa = 3.47MFFYY206 pKa = 10.72RR207 pKa = 11.84FPDD210 pKa = 3.75HH211 pKa = 6.29EE212 pKa = 4.37WSFVRR217 pKa = 11.84IGTISSRR224 pKa = 11.84YY225 pKa = 8.89KK226 pKa = 10.69DD227 pKa = 3.78CTSLLSVAHH236 pKa = 6.06TSRR239 pKa = 11.84LLGFPHH245 pKa = 7.42PAEE248 pKa = 3.82LWTWITNITLHH259 pKa = 7.93HH260 pKa = 6.24EE261 pKa = 4.17MKK263 pKa = 10.92KK264 pKa = 9.28MFNNADD270 pKa = 3.94EE271 pKa = 4.38FHH273 pKa = 7.39DD274 pKa = 4.75PYY276 pKa = 11.4SYY278 pKa = 10.85FPYY281 pKa = 9.78QIEE284 pKa = 4.08MGLSKK289 pKa = 10.66KK290 pKa = 10.25SLYY293 pKa = 8.16STRR296 pKa = 11.84EE297 pKa = 3.58NPGIHH302 pKa = 6.67AFCHH306 pKa = 6.35IIGLYY311 pKa = 7.88YY312 pKa = 10.56QNARR316 pKa = 11.84SMKK319 pKa = 9.24ARR321 pKa = 11.84WMLEE325 pKa = 3.62DD326 pKa = 4.53DD327 pKa = 4.06VNPILFNAHH336 pKa = 5.54LVGFAFTRR344 pKa = 11.84GKK346 pKa = 9.56WADD349 pKa = 3.56KK350 pKa = 8.33FWAVKK355 pKa = 9.78GHH357 pKa = 7.08IGPPAPPARR366 pKa = 11.84QPDD369 pKa = 3.7NAQQEE374 pKa = 4.44PMEE377 pKa = 4.41EE378 pKa = 4.17EE379 pKa = 3.97VDD381 pKa = 3.79EE382 pKa = 5.38EE383 pKa = 5.51DD384 pKa = 4.55IQSLNQDD391 pKa = 3.37GVDD394 pKa = 4.42AIAPPDD400 pKa = 3.77EE401 pKa = 4.57RR402 pKa = 11.84TPQAWTPYY410 pKa = 10.22LDD412 pKa = 3.33AHH414 pKa = 6.28NWTMSPAMVRR424 pKa = 11.84TAKK427 pKa = 10.36LVASGIVVEE436 pKa = 4.67RR437 pKa = 11.84DD438 pKa = 3.08YY439 pKa = 11.54TIAKK443 pKa = 9.82FSKK446 pKa = 9.6TPAFYY451 pKa = 11.28LNMDD455 pKa = 3.99
MM1 pKa = 7.97DD2 pKa = 4.47VLNLTDD8 pKa = 3.42QEE10 pKa = 4.59TNEE13 pKa = 4.17FLRR16 pKa = 11.84VRR18 pKa = 11.84AIQIEE23 pKa = 4.34KK24 pKa = 8.57TGEE27 pKa = 3.78YY28 pKa = 9.61VYY30 pKa = 10.71PWFEE34 pKa = 3.84QHH36 pKa = 6.44NSKK39 pKa = 10.7PPIKK43 pKa = 9.92IPKK46 pKa = 9.27VDD48 pKa = 3.94DD49 pKa = 3.63AAGARR54 pKa = 11.84LRR56 pKa = 11.84AITKK60 pKa = 8.99GFASGVICSVQTASAFLSWKK80 pKa = 7.7MKK82 pKa = 10.4RR83 pKa = 11.84MTARR87 pKa = 11.84VDD89 pKa = 4.25SVWKK93 pKa = 10.45SYY95 pKa = 10.71NQLIGDD101 pKa = 4.58PNADD105 pKa = 3.24ITPLSLLSVEE115 pKa = 4.7EE116 pKa = 4.29VGTPTISGNEE126 pKa = 4.05VSSSEE131 pKa = 4.93ADD133 pKa = 3.52EE134 pKa = 4.16KK135 pKa = 10.97WIIGYY140 pKa = 8.51ILCLYY145 pKa = 10.56RR146 pKa = 11.84LGRR149 pKa = 11.84VNDD152 pKa = 3.49ANYY155 pKa = 10.53RR156 pKa = 11.84GDD158 pKa = 4.52LIKK161 pKa = 10.87SMKK164 pKa = 9.75PILEE168 pKa = 4.27AQGMPNTTNLLADD181 pKa = 4.24CGKK184 pKa = 7.14TTAWCQCPVFRR195 pKa = 11.84KK196 pKa = 9.86LVAAIDD202 pKa = 3.47MFFYY206 pKa = 10.72RR207 pKa = 11.84FPDD210 pKa = 3.75HH211 pKa = 6.29EE212 pKa = 4.37WSFVRR217 pKa = 11.84IGTISSRR224 pKa = 11.84YY225 pKa = 8.89KK226 pKa = 10.69DD227 pKa = 3.78CTSLLSVAHH236 pKa = 6.06TSRR239 pKa = 11.84LLGFPHH245 pKa = 7.42PAEE248 pKa = 3.82LWTWITNITLHH259 pKa = 7.93HH260 pKa = 6.24EE261 pKa = 4.17MKK263 pKa = 10.92KK264 pKa = 9.28MFNNADD270 pKa = 3.94EE271 pKa = 4.38FHH273 pKa = 7.39DD274 pKa = 4.75PYY276 pKa = 11.4SYY278 pKa = 10.85FPYY281 pKa = 9.78QIEE284 pKa = 4.08MGLSKK289 pKa = 10.66KK290 pKa = 10.25SLYY293 pKa = 8.16STRR296 pKa = 11.84EE297 pKa = 3.58NPGIHH302 pKa = 6.67AFCHH306 pKa = 6.35IIGLYY311 pKa = 7.88YY312 pKa = 10.56QNARR316 pKa = 11.84SMKK319 pKa = 9.24ARR321 pKa = 11.84WMLEE325 pKa = 3.62DD326 pKa = 4.53DD327 pKa = 4.06VNPILFNAHH336 pKa = 5.54LVGFAFTRR344 pKa = 11.84GKK346 pKa = 9.56WADD349 pKa = 3.56KK350 pKa = 8.33FWAVKK355 pKa = 9.78GHH357 pKa = 7.08IGPPAPPARR366 pKa = 11.84QPDD369 pKa = 3.7NAQQEE374 pKa = 4.44PMEE377 pKa = 4.41EE378 pKa = 4.17EE379 pKa = 3.97VDD381 pKa = 3.79EE382 pKa = 5.38EE383 pKa = 5.51DD384 pKa = 4.55IQSLNQDD391 pKa = 3.37GVDD394 pKa = 4.42AIAPPDD400 pKa = 3.77EE401 pKa = 4.57RR402 pKa = 11.84TPQAWTPYY410 pKa = 10.22LDD412 pKa = 3.33AHH414 pKa = 6.28NWTMSPAMVRR424 pKa = 11.84TAKK427 pKa = 10.36LVASGIVVEE436 pKa = 4.67RR437 pKa = 11.84DD438 pKa = 3.08YY439 pKa = 11.54TIAKK443 pKa = 9.82FSKK446 pKa = 9.6TPAFYY451 pKa = 11.28LNMDD455 pKa = 3.99
Molecular weight: 51.68 kDa
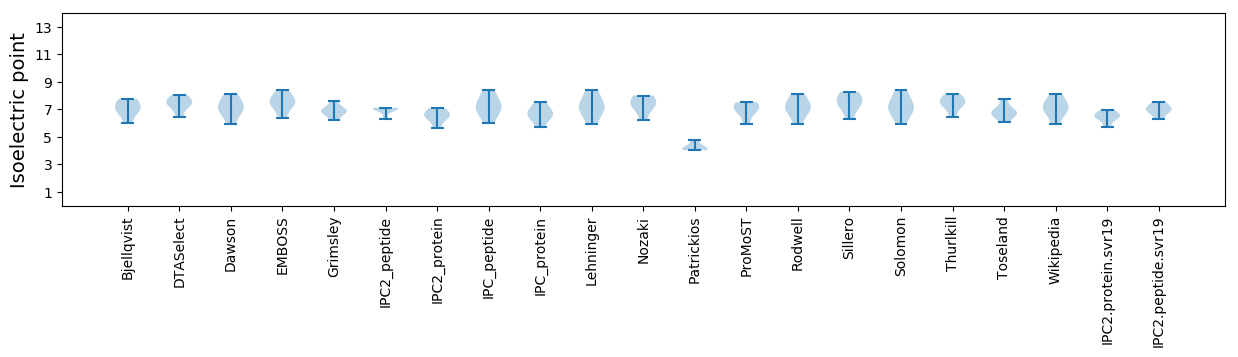
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B5KTK4|A0A0B5KTK4_9RHAB Glycoprotein OS=Wuhan House Fly Virus 1 OX=1608104 GN=G PE=4 SV=1
MM1 pKa = 7.61SNRR4 pKa = 11.84KK5 pKa = 9.17LSLGSFGKK13 pKa = 10.37AINIWKK19 pKa = 9.46PSAPSITTSSLLDD32 pKa = 3.48GVSSNWKK39 pKa = 10.12AFDD42 pKa = 4.04VFDD45 pKa = 5.89LDD47 pKa = 4.17PEE49 pKa = 4.47PTSSEE54 pKa = 4.41SATHH58 pKa = 6.59PKK60 pKa = 9.92QEE62 pKa = 4.31IVWEE66 pKa = 4.08VDD68 pKa = 2.91AQLTVSTSKK77 pKa = 10.79KK78 pKa = 7.95IAIWEE83 pKa = 4.04DD84 pKa = 3.67LIEE87 pKa = 4.47EE88 pKa = 4.44LDD90 pKa = 4.37AIIDD94 pKa = 3.79HH95 pKa = 6.75YY96 pKa = 11.21YY97 pKa = 10.76GPSNQKK103 pKa = 9.79EE104 pKa = 3.96IFIIIYY110 pKa = 9.64YY111 pKa = 10.81LLGTHH116 pKa = 7.08LKK118 pKa = 10.38PGVGVNGEE126 pKa = 4.06YY127 pKa = 10.43KK128 pKa = 10.54KK129 pKa = 10.89FVIEE133 pKa = 3.59LSKK136 pKa = 10.63RR137 pKa = 11.84IKK139 pKa = 10.25FSYY142 pKa = 10.01NFEE145 pKa = 4.58GIQRR149 pKa = 11.84EE150 pKa = 4.18AGDD153 pKa = 3.69ISWTYY158 pKa = 8.66SQSRR162 pKa = 11.84ARR164 pKa = 11.84KK165 pKa = 8.89RR166 pKa = 11.84LLIDD170 pKa = 3.67FRR172 pKa = 11.84CSITPTKK179 pKa = 10.33RR180 pKa = 11.84DD181 pKa = 3.98GIPLKK186 pKa = 10.5KK187 pKa = 10.54LYY189 pKa = 10.6DD190 pKa = 3.63FQIYY194 pKa = 9.61PNLPHH199 pKa = 6.96PSIGSLLSGNDD210 pKa = 2.88ITVLDD215 pKa = 3.83VGTNYY220 pKa = 10.52SFAKK224 pKa = 9.8KK225 pKa = 7.94QQ226 pKa = 3.2
MM1 pKa = 7.61SNRR4 pKa = 11.84KK5 pKa = 9.17LSLGSFGKK13 pKa = 10.37AINIWKK19 pKa = 9.46PSAPSITTSSLLDD32 pKa = 3.48GVSSNWKK39 pKa = 10.12AFDD42 pKa = 4.04VFDD45 pKa = 5.89LDD47 pKa = 4.17PEE49 pKa = 4.47PTSSEE54 pKa = 4.41SATHH58 pKa = 6.59PKK60 pKa = 9.92QEE62 pKa = 4.31IVWEE66 pKa = 4.08VDD68 pKa = 2.91AQLTVSTSKK77 pKa = 10.79KK78 pKa = 7.95IAIWEE83 pKa = 4.04DD84 pKa = 3.67LIEE87 pKa = 4.47EE88 pKa = 4.44LDD90 pKa = 4.37AIIDD94 pKa = 3.79HH95 pKa = 6.75YY96 pKa = 11.21YY97 pKa = 10.76GPSNQKK103 pKa = 9.79EE104 pKa = 3.96IFIIIYY110 pKa = 9.64YY111 pKa = 10.81LLGTHH116 pKa = 7.08LKK118 pKa = 10.38PGVGVNGEE126 pKa = 4.06YY127 pKa = 10.43KK128 pKa = 10.54KK129 pKa = 10.89FVIEE133 pKa = 3.59LSKK136 pKa = 10.63RR137 pKa = 11.84IKK139 pKa = 10.25FSYY142 pKa = 10.01NFEE145 pKa = 4.58GIQRR149 pKa = 11.84EE150 pKa = 4.18AGDD153 pKa = 3.69ISWTYY158 pKa = 8.66SQSRR162 pKa = 11.84ARR164 pKa = 11.84KK165 pKa = 8.89RR166 pKa = 11.84LLIDD170 pKa = 3.67FRR172 pKa = 11.84CSITPTKK179 pKa = 10.33RR180 pKa = 11.84DD181 pKa = 3.98GIPLKK186 pKa = 10.5KK187 pKa = 10.54LYY189 pKa = 10.6DD190 pKa = 3.63FQIYY194 pKa = 9.61PNLPHH199 pKa = 6.96PSIGSLLSGNDD210 pKa = 2.88ITVLDD215 pKa = 3.83VGTNYY220 pKa = 10.52SFAKK224 pKa = 9.8KK225 pKa = 7.94QQ226 pKa = 3.2
Molecular weight: 25.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3995 |
226 |
2098 |
665.8 |
75.61 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.357 ± 0.697 | 1.552 ± 0.103 |
5.682 ± 0.445 | 5.332 ± 0.122 |
4.456 ± 0.404 | 4.881 ± 0.428 |
2.804 ± 0.272 | 7.935 ± 0.547 |
5.932 ± 0.384 | 10.088 ± 0.969 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.902 ± 0.304 | 5.257 ± 0.2 |
4.906 ± 0.538 | 3.655 ± 0.187 |
4.781 ± 0.197 | 8.586 ± 0.447 |
6.558 ± 0.619 | 5.081 ± 0.432 |
1.652 ± 0.298 | 3.605 ± 0.267 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
