
Hubei hepe-like virus 2
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.59
Get precalculated fractions of proteins
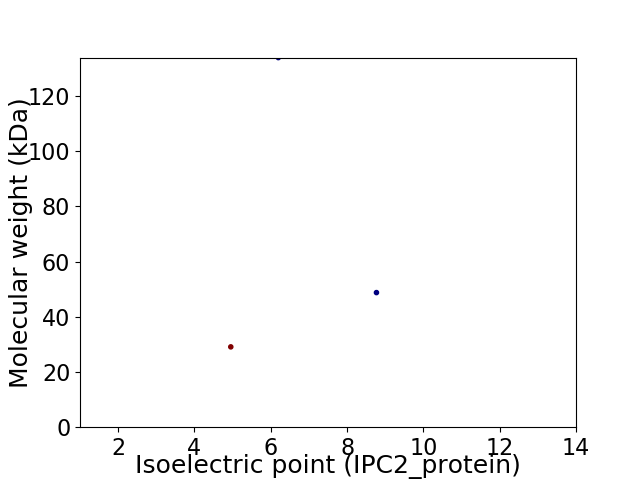
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KK91|A0A1L3KK91_9VIRU U-box domain-containing protein OS=Hubei hepe-like virus 2 OX=1922895 PE=4 SV=1
MM1 pKa = 7.34TLNLIKK7 pKa = 10.71SHH9 pKa = 6.51FSCAISKK16 pKa = 10.62SLVALPYY23 pKa = 10.35NCPCGHH29 pKa = 7.32LFDD32 pKa = 5.02AVNIFTWVNTNNTCPVSRR50 pKa = 11.84LPLLVQDD57 pKa = 3.92LTFNNTVYY65 pKa = 10.66QFLQEE70 pKa = 4.94LRR72 pKa = 11.84LTDD75 pKa = 3.69DD76 pKa = 4.05TDD78 pKa = 3.56VHH80 pKa = 7.01DD81 pKa = 4.94CATTTATEE89 pKa = 4.11ISRR92 pKa = 11.84GDD94 pKa = 3.38DD95 pKa = 3.75TIAMDD100 pKa = 5.8DD101 pKa = 3.73FTGTNDD107 pKa = 4.54LGVQTDD113 pKa = 3.83EE114 pKa = 4.74FPPLVEE120 pKa = 3.89PFVYY124 pKa = 10.44LGNEE128 pKa = 4.39VITVANLNEE137 pKa = 3.89NEE139 pKa = 3.9RR140 pKa = 11.84RR141 pKa = 11.84YY142 pKa = 10.28LGCGKK147 pKa = 10.2LVDD150 pKa = 4.18LADD153 pKa = 5.0GIFNPISTNPHH164 pKa = 6.05LEE166 pKa = 4.02QVVDD170 pKa = 4.31RR171 pKa = 11.84FNMEE175 pKa = 4.33YY176 pKa = 10.0IRR178 pKa = 11.84TLIPIRR184 pKa = 11.84ADD186 pKa = 3.09HH187 pKa = 6.61PNIVINKK194 pKa = 8.13YY195 pKa = 10.25AYY197 pKa = 7.3QTRR200 pKa = 11.84FDD202 pKa = 4.16LVATAKK208 pKa = 10.27LLCRR212 pKa = 11.84QGYY215 pKa = 8.23FVYY218 pKa = 10.76DD219 pKa = 3.63LFKK222 pKa = 10.79IGMHH226 pKa = 7.3DD227 pKa = 4.5DD228 pKa = 3.41LNQYY232 pKa = 9.22VLYY235 pKa = 8.86STPILVTGQRR245 pKa = 11.84NILSKK250 pKa = 10.67YY251 pKa = 9.85KK252 pKa = 10.68RR253 pKa = 11.84LTNN256 pKa = 3.68
MM1 pKa = 7.34TLNLIKK7 pKa = 10.71SHH9 pKa = 6.51FSCAISKK16 pKa = 10.62SLVALPYY23 pKa = 10.35NCPCGHH29 pKa = 7.32LFDD32 pKa = 5.02AVNIFTWVNTNNTCPVSRR50 pKa = 11.84LPLLVQDD57 pKa = 3.92LTFNNTVYY65 pKa = 10.66QFLQEE70 pKa = 4.94LRR72 pKa = 11.84LTDD75 pKa = 3.69DD76 pKa = 4.05TDD78 pKa = 3.56VHH80 pKa = 7.01DD81 pKa = 4.94CATTTATEE89 pKa = 4.11ISRR92 pKa = 11.84GDD94 pKa = 3.38DD95 pKa = 3.75TIAMDD100 pKa = 5.8DD101 pKa = 3.73FTGTNDD107 pKa = 4.54LGVQTDD113 pKa = 3.83EE114 pKa = 4.74FPPLVEE120 pKa = 3.89PFVYY124 pKa = 10.44LGNEE128 pKa = 4.39VITVANLNEE137 pKa = 3.89NEE139 pKa = 3.9RR140 pKa = 11.84RR141 pKa = 11.84YY142 pKa = 10.28LGCGKK147 pKa = 10.2LVDD150 pKa = 4.18LADD153 pKa = 5.0GIFNPISTNPHH164 pKa = 6.05LEE166 pKa = 4.02QVVDD170 pKa = 4.31RR171 pKa = 11.84FNMEE175 pKa = 4.33YY176 pKa = 10.0IRR178 pKa = 11.84TLIPIRR184 pKa = 11.84ADD186 pKa = 3.09HH187 pKa = 6.61PNIVINKK194 pKa = 8.13YY195 pKa = 10.25AYY197 pKa = 7.3QTRR200 pKa = 11.84FDD202 pKa = 4.16LVATAKK208 pKa = 10.27LLCRR212 pKa = 11.84QGYY215 pKa = 8.23FVYY218 pKa = 10.76DD219 pKa = 3.63LFKK222 pKa = 10.79IGMHH226 pKa = 7.3DD227 pKa = 4.5DD228 pKa = 3.41LNQYY232 pKa = 9.22VLYY235 pKa = 8.86STPILVTGQRR245 pKa = 11.84NILSKK250 pKa = 10.67YY251 pKa = 9.85KK252 pKa = 10.68RR253 pKa = 11.84LTNN256 pKa = 3.68
Molecular weight: 29.1 kDa
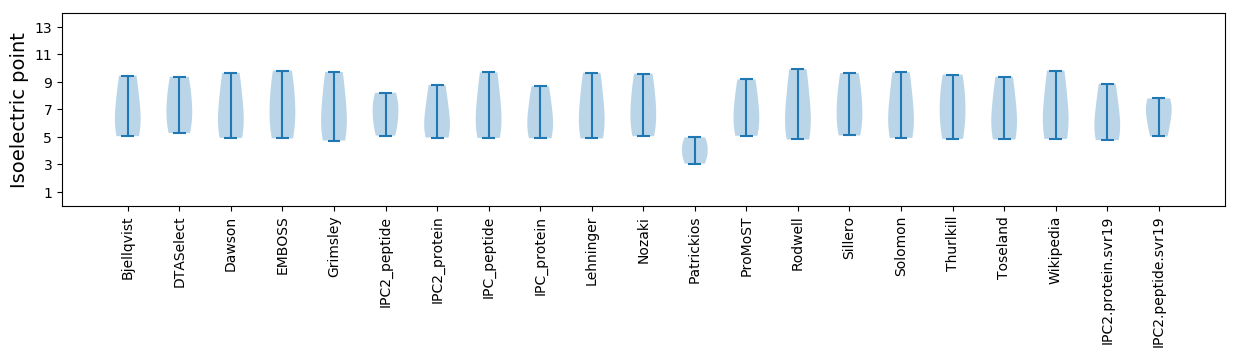
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KK26|A0A1L3KK26_9VIRU Uncharacterized protein OS=Hubei hepe-like virus 2 OX=1922895 PE=4 SV=1
MM1 pKa = 7.59VYY3 pKa = 10.24RR4 pKa = 11.84GNNRR8 pKa = 11.84NRR10 pKa = 11.84ANNNRR15 pKa = 11.84MMNNKK20 pKa = 9.48QYY22 pKa = 9.64VTVRR26 pKa = 11.84RR27 pKa = 11.84DD28 pKa = 2.93KK29 pKa = 10.94YY30 pKa = 11.17VNLISGKK37 pKa = 9.68KK38 pKa = 8.05QQSQNKK44 pKa = 8.79RR45 pKa = 11.84KK46 pKa = 9.44QPPQQRR52 pKa = 11.84KK53 pKa = 6.56QRR55 pKa = 11.84PIQRR59 pKa = 11.84PIQQVRR65 pKa = 11.84TQVFRR70 pKa = 11.84PTNRR74 pKa = 11.84KK75 pKa = 8.63PLRR78 pKa = 11.84QAARR82 pKa = 11.84KK83 pKa = 7.22TNMMNDD89 pKa = 4.24PYY91 pKa = 11.37AMCRR95 pKa = 11.84LTPFQSRR102 pKa = 11.84GLADD106 pKa = 5.57GIPDD110 pKa = 3.7GSDD113 pKa = 2.98VRR115 pKa = 11.84KK116 pKa = 9.9ILLDD120 pKa = 3.27HH121 pKa = 6.88RR122 pKa = 11.84MSNTFTFGSSGMFNILITPCIPSSIWFNSMNNDD155 pKa = 2.49STFTVNGTTYY165 pKa = 9.88PYY167 pKa = 10.83NITDD171 pKa = 4.36FNWLANTIQQPEE183 pKa = 3.94WRR185 pKa = 11.84GIPVTLLDD193 pKa = 5.03QIGKK197 pKa = 9.56FDD199 pKa = 3.99NAGSIYY205 pKa = 10.68NSGKK209 pKa = 9.8CRR211 pKa = 11.84LVTVGWSLTFIGTTVQNSGQIQVQSIGLTLGDD243 pKa = 5.23NIPNMDD249 pKa = 3.77TFSVYY254 pKa = 10.71NSQSPSNKK262 pKa = 8.67SWNNGQVMIRR272 pKa = 11.84TLNCQPLSDD281 pKa = 3.62GAMSVEE287 pKa = 4.28TKK289 pKa = 10.49VIPLRR294 pKa = 11.84SGAHH298 pKa = 5.19GVLKK302 pKa = 10.59HH303 pKa = 6.21VADD306 pKa = 4.4EE307 pKa = 4.52YY308 pKa = 10.9EE309 pKa = 4.3WTDD312 pKa = 3.28VTPNLCFIASPEE324 pKa = 3.96EE325 pKa = 3.77EE326 pKa = 4.53RR327 pKa = 11.84ICSLKK332 pKa = 10.8HH333 pKa = 5.57NEE335 pKa = 4.23NGTSEE340 pKa = 4.5NVMAHH345 pKa = 6.45WPHH348 pKa = 6.65CAAFDD353 pKa = 4.97DD354 pKa = 4.64DD355 pKa = 3.86WSTTKK360 pKa = 10.87LKK362 pKa = 9.35ITGGTTGASFVLDD375 pKa = 3.72TIYY378 pKa = 10.78CVEE381 pKa = 4.36YY382 pKa = 10.73VPSITSDD389 pKa = 3.01TYY391 pKa = 11.57AIAKK395 pKa = 9.13GSKK398 pKa = 9.1SEE400 pKa = 4.77KK401 pKa = 8.85PTLLKK406 pKa = 10.73NVNDD410 pKa = 4.21AAKK413 pKa = 9.59SLPIGSIGSAIDD425 pKa = 3.53VLKK428 pKa = 10.25TGAAIAAAVFF438 pKa = 3.7
MM1 pKa = 7.59VYY3 pKa = 10.24RR4 pKa = 11.84GNNRR8 pKa = 11.84NRR10 pKa = 11.84ANNNRR15 pKa = 11.84MMNNKK20 pKa = 9.48QYY22 pKa = 9.64VTVRR26 pKa = 11.84RR27 pKa = 11.84DD28 pKa = 2.93KK29 pKa = 10.94YY30 pKa = 11.17VNLISGKK37 pKa = 9.68KK38 pKa = 8.05QQSQNKK44 pKa = 8.79RR45 pKa = 11.84KK46 pKa = 9.44QPPQQRR52 pKa = 11.84KK53 pKa = 6.56QRR55 pKa = 11.84PIQRR59 pKa = 11.84PIQQVRR65 pKa = 11.84TQVFRR70 pKa = 11.84PTNRR74 pKa = 11.84KK75 pKa = 8.63PLRR78 pKa = 11.84QAARR82 pKa = 11.84KK83 pKa = 7.22TNMMNDD89 pKa = 4.24PYY91 pKa = 11.37AMCRR95 pKa = 11.84LTPFQSRR102 pKa = 11.84GLADD106 pKa = 5.57GIPDD110 pKa = 3.7GSDD113 pKa = 2.98VRR115 pKa = 11.84KK116 pKa = 9.9ILLDD120 pKa = 3.27HH121 pKa = 6.88RR122 pKa = 11.84MSNTFTFGSSGMFNILITPCIPSSIWFNSMNNDD155 pKa = 2.49STFTVNGTTYY165 pKa = 9.88PYY167 pKa = 10.83NITDD171 pKa = 4.36FNWLANTIQQPEE183 pKa = 3.94WRR185 pKa = 11.84GIPVTLLDD193 pKa = 5.03QIGKK197 pKa = 9.56FDD199 pKa = 3.99NAGSIYY205 pKa = 10.68NSGKK209 pKa = 9.8CRR211 pKa = 11.84LVTVGWSLTFIGTTVQNSGQIQVQSIGLTLGDD243 pKa = 5.23NIPNMDD249 pKa = 3.77TFSVYY254 pKa = 10.71NSQSPSNKK262 pKa = 8.67SWNNGQVMIRR272 pKa = 11.84TLNCQPLSDD281 pKa = 3.62GAMSVEE287 pKa = 4.28TKK289 pKa = 10.49VIPLRR294 pKa = 11.84SGAHH298 pKa = 5.19GVLKK302 pKa = 10.59HH303 pKa = 6.21VADD306 pKa = 4.4EE307 pKa = 4.52YY308 pKa = 10.9EE309 pKa = 4.3WTDD312 pKa = 3.28VTPNLCFIASPEE324 pKa = 3.96EE325 pKa = 3.77EE326 pKa = 4.53RR327 pKa = 11.84ICSLKK332 pKa = 10.8HH333 pKa = 5.57NEE335 pKa = 4.23NGTSEE340 pKa = 4.5NVMAHH345 pKa = 6.45WPHH348 pKa = 6.65CAAFDD353 pKa = 4.97DD354 pKa = 4.64DD355 pKa = 3.86WSTTKK360 pKa = 10.87LKK362 pKa = 9.35ITGGTTGASFVLDD375 pKa = 3.72TIYY378 pKa = 10.78CVEE381 pKa = 4.36YY382 pKa = 10.73VPSITSDD389 pKa = 3.01TYY391 pKa = 11.57AIAKK395 pKa = 9.13GSKK398 pKa = 9.1SEE400 pKa = 4.77KK401 pKa = 8.85PTLLKK406 pKa = 10.73NVNDD410 pKa = 4.21AAKK413 pKa = 9.59SLPIGSIGSAIDD425 pKa = 3.53VLKK428 pKa = 10.25TGAAIAAAVFF438 pKa = 3.7
Molecular weight: 48.77 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1861 |
256 |
1167 |
620.3 |
70.59 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.481 ± 0.173 | 2.311 ± 0.162 |
6.717 ± 0.558 | 4.836 ± 1.029 |
4.997 ± 0.567 | 5.535 ± 0.38 |
2.364 ± 0.366 | 7.254 ± 0.335 |
7.254 ± 1.407 | 7.63 ± 0.912 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.633 ± 0.234 | 7.2 ± 0.68 |
3.654 ± 0.808 | 3.815 ± 0.581 |
4.299 ± 0.48 | 5.857 ± 0.802 |
7.577 ± 0.577 | 5.373 ± 0.761 |
0.86 ± 0.321 | 4.352 ± 0.559 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
