
Methanohalophilus mahii (strain ATCC 35705 / DSM 5219 / SLP)
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Methanomicrobia; Methanosarcinales; Methanosarcinaceae; Methanohalophilus; Methanohalophilus mahii
Average proteome isoelectric point is 5.74
Get precalculated fractions of proteins
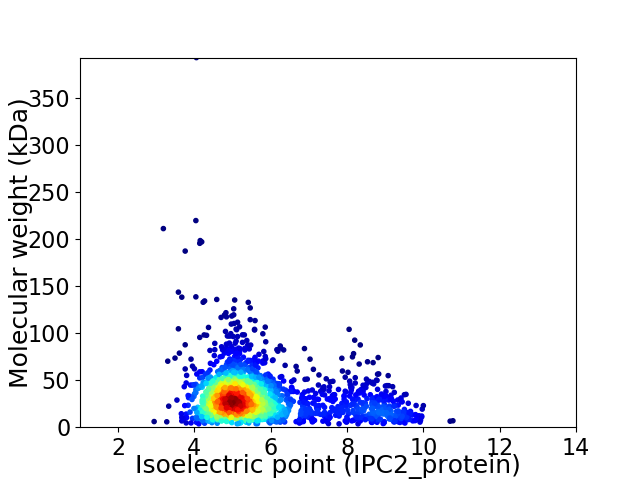
Virtual 2D-PAGE plot for 1986 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
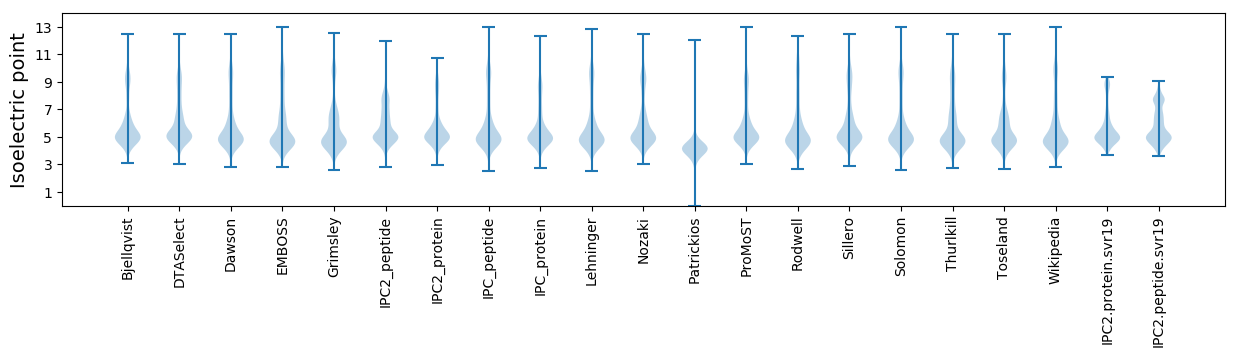
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D5E9L3|D5E9L3_METMS ORC1-type DNA replication protein OS=Methanohalophilus mahii (strain ATCC 35705 / DSM 5219 / SLP) OX=547558 GN=Mmah_0332 PE=3 SV=1
MM1 pKa = 7.08KK2 pKa = 9.93TGFKK6 pKa = 10.05ILLIGFMIIGMGCGVAAAEE25 pKa = 4.31SAPNASVTNVSSDD38 pKa = 3.27VDD40 pKa = 3.71SATIDD45 pKa = 3.72FNVNQSDD52 pKa = 3.31ASTRR56 pKa = 11.84IAYY59 pKa = 8.5GISNSLSSWSDD70 pKa = 2.78WNYY73 pKa = 10.09EE74 pKa = 3.98DD75 pKa = 4.29SNSRR79 pKa = 11.84SIVLSEE85 pKa = 4.45LNDD88 pKa = 3.24GTMYY92 pKa = 10.58YY93 pKa = 10.49YY94 pKa = 10.62SVYY97 pKa = 10.56AYY99 pKa = 10.81NGTNDD104 pKa = 3.25SFVYY108 pKa = 9.15NTTISTFNTDD118 pKa = 4.09SPTAPNASVTNVSSDD133 pKa = 3.27VDD135 pKa = 3.71SATIDD140 pKa = 3.72FNVNQSDD147 pKa = 3.31ASTRR151 pKa = 11.84IAYY154 pKa = 8.5GISNSLSSWSDD165 pKa = 2.78WNYY168 pKa = 10.09EE169 pKa = 3.98DD170 pKa = 4.29SNSRR174 pKa = 11.84SIVLSEE180 pKa = 4.0LNDD183 pKa = 3.19RR184 pKa = 11.84TMYY187 pKa = 10.27YY188 pKa = 10.33YY189 pKa = 10.61SVYY192 pKa = 10.56AYY194 pKa = 10.81NGTNDD199 pKa = 3.25SFVYY203 pKa = 9.04NTTIATLNTTSPTAPNASVTNVSSDD228 pKa = 3.27VDD230 pKa = 3.71SATIDD235 pKa = 3.72FNVNQSDD242 pKa = 3.31ASTRR246 pKa = 11.84IAYY249 pKa = 8.5GISNSLSSWSDD260 pKa = 2.78WNYY263 pKa = 10.09EE264 pKa = 3.98DD265 pKa = 4.29SNSRR269 pKa = 11.84SIVLSEE275 pKa = 4.45LNDD278 pKa = 3.24GTMYY282 pKa = 10.58YY283 pKa = 10.49YY284 pKa = 10.62SVYY287 pKa = 10.56AYY289 pKa = 10.81NGTNDD294 pKa = 3.25SFVYY298 pKa = 9.04NTTIATLNTTSPTAPSISDD317 pKa = 3.05ISSSNIGGDD326 pKa = 2.92RR327 pKa = 11.84ATINYY332 pKa = 8.29VVNQSDD338 pKa = 3.94APTRR342 pKa = 11.84IAYY345 pKa = 7.37GTSSDD350 pKa = 3.43LSSYY354 pKa = 9.51EE355 pKa = 3.44WSGWFNDD362 pKa = 3.69STVHH366 pKa = 6.92NEE368 pKa = 4.14TISNLNGSTKK378 pKa = 10.37YY379 pKa = 10.5YY380 pKa = 9.87YY381 pKa = 10.44SIYY384 pKa = 10.53AYY386 pKa = 10.47NEE388 pKa = 4.13SNNSVFYY395 pKa = 10.69NSSIEE400 pKa = 4.15SFTTISNWGDD410 pKa = 3.65RR411 pKa = 11.84IWDD414 pKa = 3.61EE415 pKa = 5.43KK416 pKa = 11.41ANQSNKK422 pKa = 7.55YY423 pKa = 7.11TWDD426 pKa = 3.22GRR428 pKa = 11.84SFSGFYY434 pKa = 10.55YY435 pKa = 10.81DD436 pKa = 6.14LDD438 pKa = 3.83TGDD441 pKa = 3.92TSEE444 pKa = 5.18SMTITIDD451 pKa = 3.43GRR453 pKa = 11.84RR454 pKa = 11.84IDD456 pKa = 5.57DD457 pKa = 3.79GDD459 pKa = 3.76LVYY462 pKa = 8.25TTKK465 pKa = 10.78PIDD468 pKa = 3.23KK469 pKa = 9.47EE470 pKa = 4.38FEE472 pKa = 3.97FGSWGEE478 pKa = 3.88YY479 pKa = 9.92QIIGFMAEE487 pKa = 4.29RR488 pKa = 11.84YY489 pKa = 8.18FAGYY493 pKa = 9.7SVNTSSDD500 pKa = 3.29VTGDD504 pKa = 3.82DD505 pKa = 3.42VSLISDD511 pKa = 4.18GVLSKK516 pKa = 11.13VLIDD520 pKa = 3.71TDD522 pKa = 4.16DD523 pKa = 4.59DD524 pKa = 4.27EE525 pKa = 5.33SVYY528 pKa = 10.96SGSSLTLEE536 pKa = 4.33EE537 pKa = 5.43GYY539 pKa = 10.69SLNIKK544 pKa = 9.82QVDD547 pKa = 3.8VNGDD551 pKa = 3.98SVWISLTKK559 pKa = 10.91DD560 pKa = 2.87GDD562 pKa = 4.09EE563 pKa = 4.24VDD565 pKa = 4.85DD566 pKa = 6.1AILSSDD572 pKa = 3.15EE573 pKa = 4.26TYY575 pKa = 11.0VYY577 pKa = 10.6EE578 pKa = 4.98KK579 pKa = 11.06DD580 pKa = 3.24LGGVDD585 pKa = 4.56DD586 pKa = 4.52VPVIAVHH593 pKa = 6.54IDD595 pKa = 3.39DD596 pKa = 4.51VFSGTEE602 pKa = 3.85TNAVFIDD609 pKa = 4.89GIFQISDD616 pKa = 3.51DD617 pKa = 4.03YY618 pKa = 12.04VEE620 pKa = 4.1VDD622 pKa = 3.23RR623 pKa = 11.84GDD625 pKa = 3.41EE626 pKa = 4.12FGEE629 pKa = 4.18MEE631 pKa = 4.4VTSLSSTEE639 pKa = 3.32IKK641 pKa = 9.92MEE643 pKa = 4.23NEE645 pKa = 3.93DD646 pKa = 5.33SISLDD651 pKa = 3.25EE652 pKa = 4.95GDD654 pKa = 5.25IIDD657 pKa = 4.97IMGKK661 pKa = 10.13LKK663 pKa = 10.86FIVADD668 pKa = 4.23DD669 pKa = 3.88NDD671 pKa = 3.93VLRR674 pKa = 11.84FAPFVDD680 pKa = 3.76MSEE683 pKa = 4.08PGTYY687 pKa = 9.25EE688 pKa = 3.51LRR690 pKa = 11.84GTIAEE695 pKa = 4.49DD696 pKa = 4.04EE697 pKa = 4.59GFSWTPLNFEE707 pKa = 4.04GFYY710 pKa = 10.87YY711 pKa = 10.65DD712 pKa = 3.07IDD714 pKa = 3.59EE715 pKa = 4.57GLRR718 pKa = 11.84SEE720 pKa = 4.52SLNVTNYY727 pKa = 10.26NGRR730 pKa = 11.84NIDD733 pKa = 4.04AGDD736 pKa = 5.11LIYY739 pKa = 10.85KK740 pKa = 6.86STPVEE745 pKa = 4.07VEE747 pKa = 3.86FEE749 pKa = 3.96YY750 pKa = 11.4GPWGKK755 pKa = 10.9YY756 pKa = 7.71NVIGFMAEE764 pKa = 3.95KK765 pKa = 10.57YY766 pKa = 9.38FAGYY770 pKa = 10.31SGDD773 pKa = 3.71TSDD776 pKa = 6.41DD777 pKa = 3.37ITDD780 pKa = 3.69DD781 pKa = 4.29TISLMSSGQLSKK793 pKa = 11.38VLIDD797 pKa = 5.23DD798 pKa = 4.84DD799 pKa = 4.88DD800 pKa = 4.28EE801 pKa = 6.5KK802 pKa = 11.42SVYY805 pKa = 9.45TGSSLILEE813 pKa = 4.41EE814 pKa = 5.82GYY816 pKa = 11.12ALNIRR821 pKa = 11.84EE822 pKa = 4.05IDD824 pKa = 3.77VNGDD828 pKa = 2.85RR829 pKa = 11.84VMIQLTQDD837 pKa = 2.9GDD839 pKa = 4.14NIEE842 pKa = 4.25SAIVSSDD849 pKa = 3.38DD850 pKa = 3.49EE851 pKa = 4.48YY852 pKa = 11.79VYY854 pKa = 11.05EE855 pKa = 5.2KK856 pKa = 11.06DD857 pKa = 4.56VGDD860 pKa = 4.28EE861 pKa = 4.1DD862 pKa = 4.38DD863 pKa = 3.8VPIIVVHH870 pKa = 6.02FNEE873 pKa = 4.5IFSGTEE879 pKa = 3.46TSAVFVEE886 pKa = 5.59GIFQISEE893 pKa = 4.24DD894 pKa = 3.9YY895 pKa = 11.56LEE897 pKa = 5.41VEE899 pKa = 5.12DD900 pKa = 5.75GDD902 pKa = 4.44DD903 pKa = 3.9FGVMTVDD910 pKa = 3.44STSGSIILEE919 pKa = 4.18NEE921 pKa = 3.87DD922 pKa = 5.49NIDD925 pKa = 3.88LDD927 pKa = 4.05EE928 pKa = 5.98GDD930 pKa = 4.7TIDD933 pKa = 4.79IMGEE937 pKa = 3.87IKK939 pKa = 10.58FKK941 pKa = 11.13VADD944 pKa = 4.12DD945 pKa = 4.05SSTVRR950 pKa = 11.84YY951 pKa = 9.26YY952 pKa = 10.49PFVEE956 pKa = 4.66VEE958 pKa = 4.22TKK960 pKa = 10.34PYY962 pKa = 10.76DD963 pKa = 3.56SLDD966 pKa = 3.18IDD968 pKa = 3.81IEE970 pKa = 4.18PTTVVEE976 pKa = 4.07GDD978 pKa = 3.91EE979 pKa = 4.04ITFTVTSRR987 pKa = 11.84GAAIRR992 pKa = 11.84DD993 pKa = 3.54ASILIHH999 pKa = 6.48GDD1001 pKa = 2.98RR1002 pKa = 11.84VGITSDD1008 pKa = 3.29EE1009 pKa = 4.38GVFEE1013 pKa = 4.35YY1014 pKa = 11.0DD1015 pKa = 3.98ADD1017 pKa = 4.09DD1018 pKa = 4.18AGKK1021 pKa = 8.46FTATAEE1027 pKa = 4.15KK1028 pKa = 10.09EE1029 pKa = 4.58GYY1031 pKa = 9.9VSSKK1035 pKa = 11.31GNFEE1039 pKa = 4.6VISPDD1044 pKa = 3.9DD1045 pKa = 3.84EE1046 pKa = 4.58SKK1048 pKa = 10.87KK1049 pKa = 10.54ISIEE1053 pKa = 3.89VSPEE1057 pKa = 3.59EE1058 pKa = 4.26VFEE1061 pKa = 4.2GTPITISVVKK1071 pKa = 10.62AIGSEE1076 pKa = 3.94PLEE1079 pKa = 4.24GAEE1082 pKa = 4.27VFFDD1086 pKa = 3.85GKK1088 pKa = 11.12SLGEE1092 pKa = 3.95TDD1094 pKa = 3.54EE1095 pKa = 5.79DD1096 pKa = 4.05GTITYY1101 pKa = 9.83NPKK1104 pKa = 10.46EE1105 pKa = 4.33PGTHH1109 pKa = 6.5KK1110 pKa = 10.51IEE1112 pKa = 4.41AKK1114 pKa = 10.04VDD1116 pKa = 3.76GYY1118 pKa = 11.64LDD1120 pKa = 3.74AEE1122 pKa = 4.56LNIKK1126 pKa = 10.1VNEE1129 pKa = 3.93LAANFEE1135 pKa = 4.29FSNLRR1140 pKa = 11.84IDD1142 pKa = 4.5PLPATAGEE1150 pKa = 4.58EE1151 pKa = 4.18FTVMLDD1157 pKa = 4.54SINTGNAEE1165 pKa = 3.57GSYY1168 pKa = 9.28TVEE1171 pKa = 4.82LSINDD1176 pKa = 3.64NVVDD1180 pKa = 3.94SQEE1183 pKa = 3.76ITLGQNNSTTIEE1195 pKa = 4.15FTSTAGEE1202 pKa = 4.17PGTYY1206 pKa = 9.43LVKK1209 pKa = 10.9AGGLSTTMDD1218 pKa = 3.56VEE1220 pKa = 4.55EE1221 pKa = 4.63GTSIVWYY1228 pKa = 10.21VLGGLVLVLGGGAAYY1243 pKa = 9.66IFATGGTAKK1252 pKa = 10.02FAEE1255 pKa = 4.46IAEE1258 pKa = 4.51SLRR1261 pKa = 11.84SRR1263 pKa = 11.84LGRR1266 pKa = 3.68
MM1 pKa = 7.08KK2 pKa = 9.93TGFKK6 pKa = 10.05ILLIGFMIIGMGCGVAAAEE25 pKa = 4.31SAPNASVTNVSSDD38 pKa = 3.27VDD40 pKa = 3.71SATIDD45 pKa = 3.72FNVNQSDD52 pKa = 3.31ASTRR56 pKa = 11.84IAYY59 pKa = 8.5GISNSLSSWSDD70 pKa = 2.78WNYY73 pKa = 10.09EE74 pKa = 3.98DD75 pKa = 4.29SNSRR79 pKa = 11.84SIVLSEE85 pKa = 4.45LNDD88 pKa = 3.24GTMYY92 pKa = 10.58YY93 pKa = 10.49YY94 pKa = 10.62SVYY97 pKa = 10.56AYY99 pKa = 10.81NGTNDD104 pKa = 3.25SFVYY108 pKa = 9.15NTTISTFNTDD118 pKa = 4.09SPTAPNASVTNVSSDD133 pKa = 3.27VDD135 pKa = 3.71SATIDD140 pKa = 3.72FNVNQSDD147 pKa = 3.31ASTRR151 pKa = 11.84IAYY154 pKa = 8.5GISNSLSSWSDD165 pKa = 2.78WNYY168 pKa = 10.09EE169 pKa = 3.98DD170 pKa = 4.29SNSRR174 pKa = 11.84SIVLSEE180 pKa = 4.0LNDD183 pKa = 3.19RR184 pKa = 11.84TMYY187 pKa = 10.27YY188 pKa = 10.33YY189 pKa = 10.61SVYY192 pKa = 10.56AYY194 pKa = 10.81NGTNDD199 pKa = 3.25SFVYY203 pKa = 9.04NTTIATLNTTSPTAPNASVTNVSSDD228 pKa = 3.27VDD230 pKa = 3.71SATIDD235 pKa = 3.72FNVNQSDD242 pKa = 3.31ASTRR246 pKa = 11.84IAYY249 pKa = 8.5GISNSLSSWSDD260 pKa = 2.78WNYY263 pKa = 10.09EE264 pKa = 3.98DD265 pKa = 4.29SNSRR269 pKa = 11.84SIVLSEE275 pKa = 4.45LNDD278 pKa = 3.24GTMYY282 pKa = 10.58YY283 pKa = 10.49YY284 pKa = 10.62SVYY287 pKa = 10.56AYY289 pKa = 10.81NGTNDD294 pKa = 3.25SFVYY298 pKa = 9.04NTTIATLNTTSPTAPSISDD317 pKa = 3.05ISSSNIGGDD326 pKa = 2.92RR327 pKa = 11.84ATINYY332 pKa = 8.29VVNQSDD338 pKa = 3.94APTRR342 pKa = 11.84IAYY345 pKa = 7.37GTSSDD350 pKa = 3.43LSSYY354 pKa = 9.51EE355 pKa = 3.44WSGWFNDD362 pKa = 3.69STVHH366 pKa = 6.92NEE368 pKa = 4.14TISNLNGSTKK378 pKa = 10.37YY379 pKa = 10.5YY380 pKa = 9.87YY381 pKa = 10.44SIYY384 pKa = 10.53AYY386 pKa = 10.47NEE388 pKa = 4.13SNNSVFYY395 pKa = 10.69NSSIEE400 pKa = 4.15SFTTISNWGDD410 pKa = 3.65RR411 pKa = 11.84IWDD414 pKa = 3.61EE415 pKa = 5.43KK416 pKa = 11.41ANQSNKK422 pKa = 7.55YY423 pKa = 7.11TWDD426 pKa = 3.22GRR428 pKa = 11.84SFSGFYY434 pKa = 10.55YY435 pKa = 10.81DD436 pKa = 6.14LDD438 pKa = 3.83TGDD441 pKa = 3.92TSEE444 pKa = 5.18SMTITIDD451 pKa = 3.43GRR453 pKa = 11.84RR454 pKa = 11.84IDD456 pKa = 5.57DD457 pKa = 3.79GDD459 pKa = 3.76LVYY462 pKa = 8.25TTKK465 pKa = 10.78PIDD468 pKa = 3.23KK469 pKa = 9.47EE470 pKa = 4.38FEE472 pKa = 3.97FGSWGEE478 pKa = 3.88YY479 pKa = 9.92QIIGFMAEE487 pKa = 4.29RR488 pKa = 11.84YY489 pKa = 8.18FAGYY493 pKa = 9.7SVNTSSDD500 pKa = 3.29VTGDD504 pKa = 3.82DD505 pKa = 3.42VSLISDD511 pKa = 4.18GVLSKK516 pKa = 11.13VLIDD520 pKa = 3.71TDD522 pKa = 4.16DD523 pKa = 4.59DD524 pKa = 4.27EE525 pKa = 5.33SVYY528 pKa = 10.96SGSSLTLEE536 pKa = 4.33EE537 pKa = 5.43GYY539 pKa = 10.69SLNIKK544 pKa = 9.82QVDD547 pKa = 3.8VNGDD551 pKa = 3.98SVWISLTKK559 pKa = 10.91DD560 pKa = 2.87GDD562 pKa = 4.09EE563 pKa = 4.24VDD565 pKa = 4.85DD566 pKa = 6.1AILSSDD572 pKa = 3.15EE573 pKa = 4.26TYY575 pKa = 11.0VYY577 pKa = 10.6EE578 pKa = 4.98KK579 pKa = 11.06DD580 pKa = 3.24LGGVDD585 pKa = 4.56DD586 pKa = 4.52VPVIAVHH593 pKa = 6.54IDD595 pKa = 3.39DD596 pKa = 4.51VFSGTEE602 pKa = 3.85TNAVFIDD609 pKa = 4.89GIFQISDD616 pKa = 3.51DD617 pKa = 4.03YY618 pKa = 12.04VEE620 pKa = 4.1VDD622 pKa = 3.23RR623 pKa = 11.84GDD625 pKa = 3.41EE626 pKa = 4.12FGEE629 pKa = 4.18MEE631 pKa = 4.4VTSLSSTEE639 pKa = 3.32IKK641 pKa = 9.92MEE643 pKa = 4.23NEE645 pKa = 3.93DD646 pKa = 5.33SISLDD651 pKa = 3.25EE652 pKa = 4.95GDD654 pKa = 5.25IIDD657 pKa = 4.97IMGKK661 pKa = 10.13LKK663 pKa = 10.86FIVADD668 pKa = 4.23DD669 pKa = 3.88NDD671 pKa = 3.93VLRR674 pKa = 11.84FAPFVDD680 pKa = 3.76MSEE683 pKa = 4.08PGTYY687 pKa = 9.25EE688 pKa = 3.51LRR690 pKa = 11.84GTIAEE695 pKa = 4.49DD696 pKa = 4.04EE697 pKa = 4.59GFSWTPLNFEE707 pKa = 4.04GFYY710 pKa = 10.87YY711 pKa = 10.65DD712 pKa = 3.07IDD714 pKa = 3.59EE715 pKa = 4.57GLRR718 pKa = 11.84SEE720 pKa = 4.52SLNVTNYY727 pKa = 10.26NGRR730 pKa = 11.84NIDD733 pKa = 4.04AGDD736 pKa = 5.11LIYY739 pKa = 10.85KK740 pKa = 6.86STPVEE745 pKa = 4.07VEE747 pKa = 3.86FEE749 pKa = 3.96YY750 pKa = 11.4GPWGKK755 pKa = 10.9YY756 pKa = 7.71NVIGFMAEE764 pKa = 3.95KK765 pKa = 10.57YY766 pKa = 9.38FAGYY770 pKa = 10.31SGDD773 pKa = 3.71TSDD776 pKa = 6.41DD777 pKa = 3.37ITDD780 pKa = 3.69DD781 pKa = 4.29TISLMSSGQLSKK793 pKa = 11.38VLIDD797 pKa = 5.23DD798 pKa = 4.84DD799 pKa = 4.88DD800 pKa = 4.28EE801 pKa = 6.5KK802 pKa = 11.42SVYY805 pKa = 9.45TGSSLILEE813 pKa = 4.41EE814 pKa = 5.82GYY816 pKa = 11.12ALNIRR821 pKa = 11.84EE822 pKa = 4.05IDD824 pKa = 3.77VNGDD828 pKa = 2.85RR829 pKa = 11.84VMIQLTQDD837 pKa = 2.9GDD839 pKa = 4.14NIEE842 pKa = 4.25SAIVSSDD849 pKa = 3.38DD850 pKa = 3.49EE851 pKa = 4.48YY852 pKa = 11.79VYY854 pKa = 11.05EE855 pKa = 5.2KK856 pKa = 11.06DD857 pKa = 4.56VGDD860 pKa = 4.28EE861 pKa = 4.1DD862 pKa = 4.38DD863 pKa = 3.8VPIIVVHH870 pKa = 6.02FNEE873 pKa = 4.5IFSGTEE879 pKa = 3.46TSAVFVEE886 pKa = 5.59GIFQISEE893 pKa = 4.24DD894 pKa = 3.9YY895 pKa = 11.56LEE897 pKa = 5.41VEE899 pKa = 5.12DD900 pKa = 5.75GDD902 pKa = 4.44DD903 pKa = 3.9FGVMTVDD910 pKa = 3.44STSGSIILEE919 pKa = 4.18NEE921 pKa = 3.87DD922 pKa = 5.49NIDD925 pKa = 3.88LDD927 pKa = 4.05EE928 pKa = 5.98GDD930 pKa = 4.7TIDD933 pKa = 4.79IMGEE937 pKa = 3.87IKK939 pKa = 10.58FKK941 pKa = 11.13VADD944 pKa = 4.12DD945 pKa = 4.05SSTVRR950 pKa = 11.84YY951 pKa = 9.26YY952 pKa = 10.49PFVEE956 pKa = 4.66VEE958 pKa = 4.22TKK960 pKa = 10.34PYY962 pKa = 10.76DD963 pKa = 3.56SLDD966 pKa = 3.18IDD968 pKa = 3.81IEE970 pKa = 4.18PTTVVEE976 pKa = 4.07GDD978 pKa = 3.91EE979 pKa = 4.04ITFTVTSRR987 pKa = 11.84GAAIRR992 pKa = 11.84DD993 pKa = 3.54ASILIHH999 pKa = 6.48GDD1001 pKa = 2.98RR1002 pKa = 11.84VGITSDD1008 pKa = 3.29EE1009 pKa = 4.38GVFEE1013 pKa = 4.35YY1014 pKa = 11.0DD1015 pKa = 3.98ADD1017 pKa = 4.09DD1018 pKa = 4.18AGKK1021 pKa = 8.46FTATAEE1027 pKa = 4.15KK1028 pKa = 10.09EE1029 pKa = 4.58GYY1031 pKa = 9.9VSSKK1035 pKa = 11.31GNFEE1039 pKa = 4.6VISPDD1044 pKa = 3.9DD1045 pKa = 3.84EE1046 pKa = 4.58SKK1048 pKa = 10.87KK1049 pKa = 10.54ISIEE1053 pKa = 3.89VSPEE1057 pKa = 3.59EE1058 pKa = 4.26VFEE1061 pKa = 4.2GTPITISVVKK1071 pKa = 10.62AIGSEE1076 pKa = 3.94PLEE1079 pKa = 4.24GAEE1082 pKa = 4.27VFFDD1086 pKa = 3.85GKK1088 pKa = 11.12SLGEE1092 pKa = 3.95TDD1094 pKa = 3.54EE1095 pKa = 5.79DD1096 pKa = 4.05GTITYY1101 pKa = 9.83NPKK1104 pKa = 10.46EE1105 pKa = 4.33PGTHH1109 pKa = 6.5KK1110 pKa = 10.51IEE1112 pKa = 4.41AKK1114 pKa = 10.04VDD1116 pKa = 3.76GYY1118 pKa = 11.64LDD1120 pKa = 3.74AEE1122 pKa = 4.56LNIKK1126 pKa = 10.1VNEE1129 pKa = 3.93LAANFEE1135 pKa = 4.29FSNLRR1140 pKa = 11.84IDD1142 pKa = 4.5PLPATAGEE1150 pKa = 4.58EE1151 pKa = 4.18FTVMLDD1157 pKa = 4.54SINTGNAEE1165 pKa = 3.57GSYY1168 pKa = 9.28TVEE1171 pKa = 4.82LSINDD1176 pKa = 3.64NVVDD1180 pKa = 3.94SQEE1183 pKa = 3.76ITLGQNNSTTIEE1195 pKa = 4.15FTSTAGEE1202 pKa = 4.17PGTYY1206 pKa = 9.43LVKK1209 pKa = 10.9AGGLSTTMDD1218 pKa = 3.56VEE1220 pKa = 4.55EE1221 pKa = 4.63GTSIVWYY1228 pKa = 10.21VLGGLVLVLGGGAAYY1243 pKa = 9.66IFATGGTAKK1252 pKa = 10.02FAEE1255 pKa = 4.46IAEE1258 pKa = 4.51SLRR1261 pKa = 11.84SRR1263 pKa = 11.84LGRR1266 pKa = 3.68
Molecular weight: 138.46 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D5EC60|D5EC60_METMS Uncharacterized protein OS=Methanohalophilus mahii (strain ATCC 35705 / DSM 5219 / SLP) OX=547558 GN=Mmah_1257 PE=4 SV=1
MM1 pKa = 7.52DD2 pKa = 3.41QVRR5 pKa = 11.84QVSHH9 pKa = 4.82NTKK12 pKa = 8.26GQKK15 pKa = 8.66MRR17 pKa = 11.84LAKK20 pKa = 10.61AHH22 pKa = 5.45VQNQRR27 pKa = 11.84VPTWAIIKK35 pKa = 9.51SNRR38 pKa = 11.84KK39 pKa = 8.84VVSHH43 pKa = 6.56PKK45 pKa = 8.36RR46 pKa = 11.84RR47 pKa = 11.84HH48 pKa = 3.55WRR50 pKa = 11.84RR51 pKa = 11.84NSLKK55 pKa = 10.72VKK57 pKa = 10.47
MM1 pKa = 7.52DD2 pKa = 3.41QVRR5 pKa = 11.84QVSHH9 pKa = 4.82NTKK12 pKa = 8.26GQKK15 pKa = 8.66MRR17 pKa = 11.84LAKK20 pKa = 10.61AHH22 pKa = 5.45VQNQRR27 pKa = 11.84VPTWAIIKK35 pKa = 9.51SNRR38 pKa = 11.84KK39 pKa = 8.84VVSHH43 pKa = 6.56PKK45 pKa = 8.36RR46 pKa = 11.84RR47 pKa = 11.84HH48 pKa = 3.55WRR50 pKa = 11.84RR51 pKa = 11.84NSLKK55 pKa = 10.72VKK57 pKa = 10.47
Molecular weight: 6.85 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
589724 |
31 |
3593 |
296.9 |
33.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.122 ± 0.061 | 1.304 ± 0.032 |
6.221 ± 0.053 | 7.659 ± 0.06 |
3.784 ± 0.041 | 7.384 ± 0.053 |
1.942 ± 0.024 | 8.012 ± 0.048 |
6.112 ± 0.061 | 8.762 ± 0.068 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.927 ± 0.03 | 4.462 ± 0.066 |
3.888 ± 0.029 | 2.757 ± 0.028 |
4.435 ± 0.056 | 6.457 ± 0.057 |
5.306 ± 0.053 | 7.196 ± 0.047 |
0.845 ± 0.018 | 3.423 ± 0.041 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
