
narna-like virus 6
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.83
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A1L3KIM6|A0A1L3KIM6_9VIRU RNA-dependent RNA polymerase OS=narna-like virus 6 OX=1923780 PE=4 SV=1
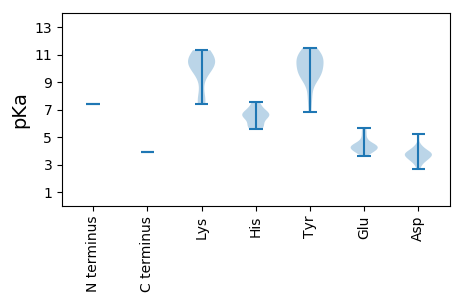
MM1 pKa = 7.38LNVYY5 pKa = 10.03HH6 pKa = 6.6FVYY9 pKa = 10.03RR10 pKa = 11.84QYY12 pKa = 11.38GFHH15 pKa = 6.67FRR17 pKa = 11.84PSRR20 pKa = 11.84EE21 pKa = 3.91DD22 pKa = 3.66LIFWEE27 pKa = 4.39WLGATNRR34 pKa = 11.84LEE36 pKa = 4.91TYY38 pKa = 10.25LKK40 pKa = 10.13WKK42 pKa = 7.39TANMLSRR49 pKa = 11.84ALGSSEE55 pKa = 5.65LVPCPLDD62 pKa = 3.72LSHH65 pKa = 7.25IPPHH69 pKa = 5.92FDD71 pKa = 3.3YY72 pKa = 11.49YY73 pKa = 11.03FGADD77 pKa = 3.14RR78 pKa = 11.84LGLLRR83 pKa = 11.84TDD85 pKa = 3.61QGFHH89 pKa = 6.65RR90 pKa = 11.84MKK92 pKa = 10.62LHH94 pKa = 6.64RR95 pKa = 11.84KK96 pKa = 7.61STDD99 pKa = 2.9RR100 pKa = 11.84YY101 pKa = 10.59ASFCFSLYY109 pKa = 9.35QSKK112 pKa = 10.18AASLPAEE119 pKa = 4.2EE120 pKa = 3.94WRR122 pKa = 11.84VRR124 pKa = 11.84SEE126 pKa = 3.62VVAAMRR132 pKa = 11.84RR133 pKa = 11.84LTEE136 pKa = 4.24KK137 pKa = 10.39PDD139 pKa = 3.74PLVPKK144 pKa = 10.15ILTNGARR151 pKa = 11.84LDD153 pKa = 3.76YY154 pKa = 11.0DD155 pKa = 3.89LLEE158 pKa = 4.54EE159 pKa = 4.32AVHH162 pKa = 5.71RR163 pKa = 11.84TIEE166 pKa = 4.42LFSPCNRR173 pKa = 11.84GDD175 pKa = 3.26RR176 pKa = 11.84LARR179 pKa = 11.84RR180 pKa = 11.84RR181 pKa = 11.84LPSRR185 pKa = 11.84GAVLGSSRR193 pKa = 11.84SEE195 pKa = 3.93GGALGQIARR204 pKa = 11.84LGHH207 pKa = 5.88GCDD210 pKa = 3.48FFVPDD215 pKa = 3.98RR216 pKa = 11.84YY217 pKa = 10.86LIGFYY222 pKa = 10.0EE223 pKa = 5.24GPAGAVEE230 pKa = 4.38VRR232 pKa = 11.84SSCEE236 pKa = 3.86PEE238 pKa = 4.03DD239 pKa = 3.77VAWAYY244 pKa = 11.04DD245 pKa = 4.18FFGHH249 pKa = 6.82WGMSHH254 pKa = 7.56DD255 pKa = 4.22VPCWPVGLPEE265 pKa = 4.15PFKK268 pKa = 11.31VRR270 pKa = 11.84VITKK274 pKa = 9.96GYY276 pKa = 9.3APSYY280 pKa = 8.19NHH282 pKa = 6.4ARR284 pKa = 11.84AYY286 pKa = 9.87QPRR289 pKa = 11.84LWRR292 pKa = 11.84SLQEE296 pKa = 4.01HH297 pKa = 5.59NVFRR301 pKa = 11.84LTGEE305 pKa = 4.19MVTGSAIQSFLSEE318 pKa = 4.62CPVDD322 pKa = 3.71SGAFFLSGDD331 pKa = 3.75YY332 pKa = 10.87KK333 pKa = 10.82EE334 pKa = 4.85ATDD337 pKa = 5.24HH338 pKa = 6.59IPSEE342 pKa = 4.4IANLMCMRR350 pKa = 11.84FCDD353 pKa = 3.48RR354 pKa = 11.84LGIPLEE360 pKa = 4.15VVGRR364 pKa = 11.84IARR367 pKa = 11.84ALTGHH372 pKa = 7.43ILPTPEE378 pKa = 5.12LVDD381 pKa = 3.37PTMGLGSQGCMVRR394 pKa = 11.84RR395 pKa = 11.84QKK397 pKa = 10.57KK398 pKa = 7.7GQLMGSPISFPFLCLYY414 pKa = 10.9NLVLQILFHH423 pKa = 6.53FVAEE427 pKa = 4.64GEE429 pKa = 4.35WIRR432 pKa = 11.84DD433 pKa = 3.36LSGVRR438 pKa = 11.84NLINGDD444 pKa = 3.89DD445 pKa = 3.8LLSYY449 pKa = 10.51LRR451 pKa = 11.84EE452 pKa = 4.0RR453 pKa = 11.84GLYY456 pKa = 9.65RR457 pKa = 11.84IWEE460 pKa = 4.28EE461 pKa = 3.87VVEE464 pKa = 4.26WGGLKK469 pKa = 10.39PSVGKK474 pKa = 8.85TIVSQRR480 pKa = 11.84YY481 pKa = 6.82LTINSKK487 pKa = 10.14LFRR490 pKa = 11.84VDD492 pKa = 3.38SAVDD496 pKa = 3.25QHH498 pKa = 5.83GTPYY502 pKa = 9.36WRR504 pKa = 11.84NRR506 pKa = 11.84HH507 pKa = 5.62LPHH510 pKa = 6.3VQLQLAIGSMKK521 pKa = 10.28SGHH524 pKa = 6.48IDD526 pKa = 3.07EE527 pKa = 5.55DD528 pKa = 3.92SWVLSSTSPRR538 pKa = 11.84SRR540 pKa = 11.84SRR542 pKa = 11.84MWQEE546 pKa = 4.41FLDD549 pKa = 4.06SCPDD553 pKa = 3.45KK554 pKa = 11.32DD555 pKa = 4.09RR556 pKa = 11.84AWSFLFSANRR566 pKa = 11.84GLLRR570 pKa = 11.84GLMDD574 pKa = 4.94KK575 pKa = 11.18YY576 pKa = 9.57PTCTLCLPTEE586 pKa = 4.3AGGLGFPLPPIDD598 pKa = 4.2SRR600 pKa = 11.84WYY602 pKa = 7.72VQRR605 pKa = 11.84APRR608 pKa = 11.84SVDD611 pKa = 2.82RR612 pKa = 11.84MKK614 pKa = 11.24ARR616 pKa = 11.84LLLSAGLPSHH626 pKa = 6.96DD627 pKa = 3.72QLRR630 pKa = 11.84SRR632 pKa = 11.84WLSSLASEE640 pKa = 5.43DD641 pKa = 3.84PRR643 pKa = 11.84TADD646 pKa = 3.37ALLFGDD652 pKa = 4.4LLRR655 pKa = 11.84YY656 pKa = 9.03QKK658 pKa = 9.69STGCPLVLEE667 pKa = 4.54TLDD670 pKa = 3.58EE671 pKa = 4.27QVGKK675 pKa = 10.61EE676 pKa = 4.07KK677 pKa = 10.77DD678 pKa = 3.82LPPPILACPLAGGVIDD694 pKa = 4.39PLEE697 pKa = 4.07EE698 pKa = 4.38HH699 pKa = 6.94LLLASQRR706 pKa = 11.84RR707 pKa = 11.84VIRR710 pKa = 11.84DD711 pKa = 2.67IDD713 pKa = 3.65RR714 pKa = 11.84AVRR717 pKa = 11.84AYY719 pKa = 10.35SRR721 pKa = 11.84SGRR724 pKa = 11.84GPWTQCEE731 pKa = 3.75ISDD734 pKa = 3.66FLRR737 pKa = 11.84RR738 pKa = 11.84FAWRR742 pKa = 11.84RR743 pKa = 11.84KK744 pKa = 7.47YY745 pKa = 10.74GRR747 pKa = 11.84YY748 pKa = 9.5LDD750 pKa = 3.93RR751 pKa = 11.84TCGFF755 pKa = 3.9
MM1 pKa = 7.38LNVYY5 pKa = 10.03HH6 pKa = 6.6FVYY9 pKa = 10.03RR10 pKa = 11.84QYY12 pKa = 11.38GFHH15 pKa = 6.67FRR17 pKa = 11.84PSRR20 pKa = 11.84EE21 pKa = 3.91DD22 pKa = 3.66LIFWEE27 pKa = 4.39WLGATNRR34 pKa = 11.84LEE36 pKa = 4.91TYY38 pKa = 10.25LKK40 pKa = 10.13WKK42 pKa = 7.39TANMLSRR49 pKa = 11.84ALGSSEE55 pKa = 5.65LVPCPLDD62 pKa = 3.72LSHH65 pKa = 7.25IPPHH69 pKa = 5.92FDD71 pKa = 3.3YY72 pKa = 11.49YY73 pKa = 11.03FGADD77 pKa = 3.14RR78 pKa = 11.84LGLLRR83 pKa = 11.84TDD85 pKa = 3.61QGFHH89 pKa = 6.65RR90 pKa = 11.84MKK92 pKa = 10.62LHH94 pKa = 6.64RR95 pKa = 11.84KK96 pKa = 7.61STDD99 pKa = 2.9RR100 pKa = 11.84YY101 pKa = 10.59ASFCFSLYY109 pKa = 9.35QSKK112 pKa = 10.18AASLPAEE119 pKa = 4.2EE120 pKa = 3.94WRR122 pKa = 11.84VRR124 pKa = 11.84SEE126 pKa = 3.62VVAAMRR132 pKa = 11.84RR133 pKa = 11.84LTEE136 pKa = 4.24KK137 pKa = 10.39PDD139 pKa = 3.74PLVPKK144 pKa = 10.15ILTNGARR151 pKa = 11.84LDD153 pKa = 3.76YY154 pKa = 11.0DD155 pKa = 3.89LLEE158 pKa = 4.54EE159 pKa = 4.32AVHH162 pKa = 5.71RR163 pKa = 11.84TIEE166 pKa = 4.42LFSPCNRR173 pKa = 11.84GDD175 pKa = 3.26RR176 pKa = 11.84LARR179 pKa = 11.84RR180 pKa = 11.84RR181 pKa = 11.84LPSRR185 pKa = 11.84GAVLGSSRR193 pKa = 11.84SEE195 pKa = 3.93GGALGQIARR204 pKa = 11.84LGHH207 pKa = 5.88GCDD210 pKa = 3.48FFVPDD215 pKa = 3.98RR216 pKa = 11.84YY217 pKa = 10.86LIGFYY222 pKa = 10.0EE223 pKa = 5.24GPAGAVEE230 pKa = 4.38VRR232 pKa = 11.84SSCEE236 pKa = 3.86PEE238 pKa = 4.03DD239 pKa = 3.77VAWAYY244 pKa = 11.04DD245 pKa = 4.18FFGHH249 pKa = 6.82WGMSHH254 pKa = 7.56DD255 pKa = 4.22VPCWPVGLPEE265 pKa = 4.15PFKK268 pKa = 11.31VRR270 pKa = 11.84VITKK274 pKa = 9.96GYY276 pKa = 9.3APSYY280 pKa = 8.19NHH282 pKa = 6.4ARR284 pKa = 11.84AYY286 pKa = 9.87QPRR289 pKa = 11.84LWRR292 pKa = 11.84SLQEE296 pKa = 4.01HH297 pKa = 5.59NVFRR301 pKa = 11.84LTGEE305 pKa = 4.19MVTGSAIQSFLSEE318 pKa = 4.62CPVDD322 pKa = 3.71SGAFFLSGDD331 pKa = 3.75YY332 pKa = 10.87KK333 pKa = 10.82EE334 pKa = 4.85ATDD337 pKa = 5.24HH338 pKa = 6.59IPSEE342 pKa = 4.4IANLMCMRR350 pKa = 11.84FCDD353 pKa = 3.48RR354 pKa = 11.84LGIPLEE360 pKa = 4.15VVGRR364 pKa = 11.84IARR367 pKa = 11.84ALTGHH372 pKa = 7.43ILPTPEE378 pKa = 5.12LVDD381 pKa = 3.37PTMGLGSQGCMVRR394 pKa = 11.84RR395 pKa = 11.84QKK397 pKa = 10.57KK398 pKa = 7.7GQLMGSPISFPFLCLYY414 pKa = 10.9NLVLQILFHH423 pKa = 6.53FVAEE427 pKa = 4.64GEE429 pKa = 4.35WIRR432 pKa = 11.84DD433 pKa = 3.36LSGVRR438 pKa = 11.84NLINGDD444 pKa = 3.89DD445 pKa = 3.8LLSYY449 pKa = 10.51LRR451 pKa = 11.84EE452 pKa = 4.0RR453 pKa = 11.84GLYY456 pKa = 9.65RR457 pKa = 11.84IWEE460 pKa = 4.28EE461 pKa = 3.87VVEE464 pKa = 4.26WGGLKK469 pKa = 10.39PSVGKK474 pKa = 8.85TIVSQRR480 pKa = 11.84YY481 pKa = 6.82LTINSKK487 pKa = 10.14LFRR490 pKa = 11.84VDD492 pKa = 3.38SAVDD496 pKa = 3.25QHH498 pKa = 5.83GTPYY502 pKa = 9.36WRR504 pKa = 11.84NRR506 pKa = 11.84HH507 pKa = 5.62LPHH510 pKa = 6.3VQLQLAIGSMKK521 pKa = 10.28SGHH524 pKa = 6.48IDD526 pKa = 3.07EE527 pKa = 5.55DD528 pKa = 3.92SWVLSSTSPRR538 pKa = 11.84SRR540 pKa = 11.84SRR542 pKa = 11.84MWQEE546 pKa = 4.41FLDD549 pKa = 4.06SCPDD553 pKa = 3.45KK554 pKa = 11.32DD555 pKa = 4.09RR556 pKa = 11.84AWSFLFSANRR566 pKa = 11.84GLLRR570 pKa = 11.84GLMDD574 pKa = 4.94KK575 pKa = 11.18YY576 pKa = 9.57PTCTLCLPTEE586 pKa = 4.3AGGLGFPLPPIDD598 pKa = 4.2SRR600 pKa = 11.84WYY602 pKa = 7.72VQRR605 pKa = 11.84APRR608 pKa = 11.84SVDD611 pKa = 2.82RR612 pKa = 11.84MKK614 pKa = 11.24ARR616 pKa = 11.84LLLSAGLPSHH626 pKa = 6.96DD627 pKa = 3.72QLRR630 pKa = 11.84SRR632 pKa = 11.84WLSSLASEE640 pKa = 5.43DD641 pKa = 3.84PRR643 pKa = 11.84TADD646 pKa = 3.37ALLFGDD652 pKa = 4.4LLRR655 pKa = 11.84YY656 pKa = 9.03QKK658 pKa = 9.69STGCPLVLEE667 pKa = 4.54TLDD670 pKa = 3.58EE671 pKa = 4.27QVGKK675 pKa = 10.61EE676 pKa = 4.07KK677 pKa = 10.77DD678 pKa = 3.82LPPPILACPLAGGVIDD694 pKa = 4.39PLEE697 pKa = 4.07EE698 pKa = 4.38HH699 pKa = 6.94LLLASQRR706 pKa = 11.84RR707 pKa = 11.84VIRR710 pKa = 11.84DD711 pKa = 2.67IDD713 pKa = 3.65RR714 pKa = 11.84AVRR717 pKa = 11.84AYY719 pKa = 10.35SRR721 pKa = 11.84SGRR724 pKa = 11.84GPWTQCEE731 pKa = 3.75ISDD734 pKa = 3.66FLRR737 pKa = 11.84RR738 pKa = 11.84FAWRR742 pKa = 11.84RR743 pKa = 11.84KK744 pKa = 7.47YY745 pKa = 10.74GRR747 pKa = 11.84YY748 pKa = 9.5LDD750 pKa = 3.93RR751 pKa = 11.84TCGFF755 pKa = 3.9
Molecular weight: 86.0 kDa
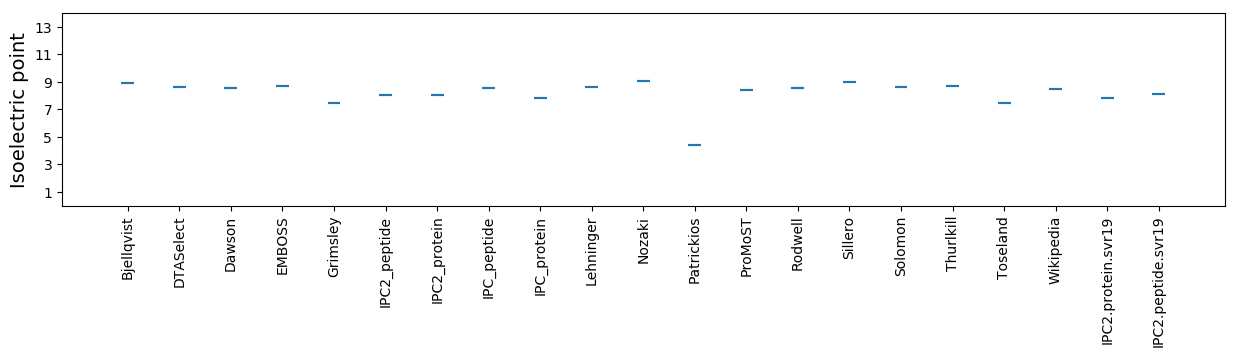
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KIM6|A0A1L3KIM6_9VIRU RNA-dependent RNA polymerase OS=narna-like virus 6 OX=1923780 PE=4 SV=1
MM1 pKa = 7.38LNVYY5 pKa = 10.03HH6 pKa = 6.6FVYY9 pKa = 10.03RR10 pKa = 11.84QYY12 pKa = 11.38GFHH15 pKa = 6.67FRR17 pKa = 11.84PSRR20 pKa = 11.84EE21 pKa = 3.91DD22 pKa = 3.66LIFWEE27 pKa = 4.39WLGATNRR34 pKa = 11.84LEE36 pKa = 4.91TYY38 pKa = 10.25LKK40 pKa = 10.13WKK42 pKa = 7.39TANMLSRR49 pKa = 11.84ALGSSEE55 pKa = 5.65LVPCPLDD62 pKa = 3.72LSHH65 pKa = 7.25IPPHH69 pKa = 5.92FDD71 pKa = 3.3YY72 pKa = 11.49YY73 pKa = 11.03FGADD77 pKa = 3.14RR78 pKa = 11.84LGLLRR83 pKa = 11.84TDD85 pKa = 3.61QGFHH89 pKa = 6.65RR90 pKa = 11.84MKK92 pKa = 10.62LHH94 pKa = 6.64RR95 pKa = 11.84KK96 pKa = 7.61STDD99 pKa = 2.9RR100 pKa = 11.84YY101 pKa = 10.59ASFCFSLYY109 pKa = 9.35QSKK112 pKa = 10.18AASLPAEE119 pKa = 4.2EE120 pKa = 3.94WRR122 pKa = 11.84VRR124 pKa = 11.84SEE126 pKa = 3.62VVAAMRR132 pKa = 11.84RR133 pKa = 11.84LTEE136 pKa = 4.24KK137 pKa = 10.39PDD139 pKa = 3.74PLVPKK144 pKa = 10.15ILTNGARR151 pKa = 11.84LDD153 pKa = 3.76YY154 pKa = 11.0DD155 pKa = 3.89LLEE158 pKa = 4.54EE159 pKa = 4.32AVHH162 pKa = 5.71RR163 pKa = 11.84TIEE166 pKa = 4.42LFSPCNRR173 pKa = 11.84GDD175 pKa = 3.26RR176 pKa = 11.84LARR179 pKa = 11.84RR180 pKa = 11.84RR181 pKa = 11.84LPSRR185 pKa = 11.84GAVLGSSRR193 pKa = 11.84SEE195 pKa = 3.93GGALGQIARR204 pKa = 11.84LGHH207 pKa = 5.88GCDD210 pKa = 3.48FFVPDD215 pKa = 3.98RR216 pKa = 11.84YY217 pKa = 10.86LIGFYY222 pKa = 10.0EE223 pKa = 5.24GPAGAVEE230 pKa = 4.38VRR232 pKa = 11.84SSCEE236 pKa = 3.86PEE238 pKa = 4.03DD239 pKa = 3.77VAWAYY244 pKa = 11.04DD245 pKa = 4.18FFGHH249 pKa = 6.82WGMSHH254 pKa = 7.56DD255 pKa = 4.22VPCWPVGLPEE265 pKa = 4.15PFKK268 pKa = 11.31VRR270 pKa = 11.84VITKK274 pKa = 9.96GYY276 pKa = 9.3APSYY280 pKa = 8.19NHH282 pKa = 6.4ARR284 pKa = 11.84AYY286 pKa = 9.87QPRR289 pKa = 11.84LWRR292 pKa = 11.84SLQEE296 pKa = 4.01HH297 pKa = 5.59NVFRR301 pKa = 11.84LTGEE305 pKa = 4.19MVTGSAIQSFLSEE318 pKa = 4.62CPVDD322 pKa = 3.71SGAFFLSGDD331 pKa = 3.75YY332 pKa = 10.87KK333 pKa = 10.82EE334 pKa = 4.85ATDD337 pKa = 5.24HH338 pKa = 6.59IPSEE342 pKa = 4.4IANLMCMRR350 pKa = 11.84FCDD353 pKa = 3.48RR354 pKa = 11.84LGIPLEE360 pKa = 4.15VVGRR364 pKa = 11.84IARR367 pKa = 11.84ALTGHH372 pKa = 7.43ILPTPEE378 pKa = 5.12LVDD381 pKa = 3.37PTMGLGSQGCMVRR394 pKa = 11.84RR395 pKa = 11.84QKK397 pKa = 10.57KK398 pKa = 7.7GQLMGSPISFPFLCLYY414 pKa = 10.9NLVLQILFHH423 pKa = 6.53FVAEE427 pKa = 4.64GEE429 pKa = 4.35WIRR432 pKa = 11.84DD433 pKa = 3.36LSGVRR438 pKa = 11.84NLINGDD444 pKa = 3.89DD445 pKa = 3.8LLSYY449 pKa = 10.51LRR451 pKa = 11.84EE452 pKa = 4.0RR453 pKa = 11.84GLYY456 pKa = 9.65RR457 pKa = 11.84IWEE460 pKa = 4.28EE461 pKa = 3.87VVEE464 pKa = 4.26WGGLKK469 pKa = 10.39PSVGKK474 pKa = 8.85TIVSQRR480 pKa = 11.84YY481 pKa = 6.82LTINSKK487 pKa = 10.14LFRR490 pKa = 11.84VDD492 pKa = 3.38SAVDD496 pKa = 3.25QHH498 pKa = 5.83GTPYY502 pKa = 9.36WRR504 pKa = 11.84NRR506 pKa = 11.84HH507 pKa = 5.62LPHH510 pKa = 6.3VQLQLAIGSMKK521 pKa = 10.28SGHH524 pKa = 6.48IDD526 pKa = 3.07EE527 pKa = 5.55DD528 pKa = 3.92SWVLSSTSPRR538 pKa = 11.84SRR540 pKa = 11.84SRR542 pKa = 11.84MWQEE546 pKa = 4.41FLDD549 pKa = 4.06SCPDD553 pKa = 3.45KK554 pKa = 11.32DD555 pKa = 4.09RR556 pKa = 11.84AWSFLFSANRR566 pKa = 11.84GLLRR570 pKa = 11.84GLMDD574 pKa = 4.94KK575 pKa = 11.18YY576 pKa = 9.57PTCTLCLPTEE586 pKa = 4.3AGGLGFPLPPIDD598 pKa = 4.2SRR600 pKa = 11.84WYY602 pKa = 7.72VQRR605 pKa = 11.84APRR608 pKa = 11.84SVDD611 pKa = 2.82RR612 pKa = 11.84MKK614 pKa = 11.24ARR616 pKa = 11.84LLLSAGLPSHH626 pKa = 6.96DD627 pKa = 3.72QLRR630 pKa = 11.84SRR632 pKa = 11.84WLSSLASEE640 pKa = 5.43DD641 pKa = 3.84PRR643 pKa = 11.84TADD646 pKa = 3.37ALLFGDD652 pKa = 4.4LLRR655 pKa = 11.84YY656 pKa = 9.03QKK658 pKa = 9.69STGCPLVLEE667 pKa = 4.54TLDD670 pKa = 3.58EE671 pKa = 4.27QVGKK675 pKa = 10.61EE676 pKa = 4.07KK677 pKa = 10.77DD678 pKa = 3.82LPPPILACPLAGGVIDD694 pKa = 4.39PLEE697 pKa = 4.07EE698 pKa = 4.38HH699 pKa = 6.94LLLASQRR706 pKa = 11.84RR707 pKa = 11.84VIRR710 pKa = 11.84DD711 pKa = 2.67IDD713 pKa = 3.65RR714 pKa = 11.84AVRR717 pKa = 11.84AYY719 pKa = 10.35SRR721 pKa = 11.84SGRR724 pKa = 11.84GPWTQCEE731 pKa = 3.75ISDD734 pKa = 3.66FLRR737 pKa = 11.84RR738 pKa = 11.84FAWRR742 pKa = 11.84RR743 pKa = 11.84KK744 pKa = 7.47YY745 pKa = 10.74GRR747 pKa = 11.84YY748 pKa = 9.5LDD750 pKa = 3.93RR751 pKa = 11.84TCGFF755 pKa = 3.9
MM1 pKa = 7.38LNVYY5 pKa = 10.03HH6 pKa = 6.6FVYY9 pKa = 10.03RR10 pKa = 11.84QYY12 pKa = 11.38GFHH15 pKa = 6.67FRR17 pKa = 11.84PSRR20 pKa = 11.84EE21 pKa = 3.91DD22 pKa = 3.66LIFWEE27 pKa = 4.39WLGATNRR34 pKa = 11.84LEE36 pKa = 4.91TYY38 pKa = 10.25LKK40 pKa = 10.13WKK42 pKa = 7.39TANMLSRR49 pKa = 11.84ALGSSEE55 pKa = 5.65LVPCPLDD62 pKa = 3.72LSHH65 pKa = 7.25IPPHH69 pKa = 5.92FDD71 pKa = 3.3YY72 pKa = 11.49YY73 pKa = 11.03FGADD77 pKa = 3.14RR78 pKa = 11.84LGLLRR83 pKa = 11.84TDD85 pKa = 3.61QGFHH89 pKa = 6.65RR90 pKa = 11.84MKK92 pKa = 10.62LHH94 pKa = 6.64RR95 pKa = 11.84KK96 pKa = 7.61STDD99 pKa = 2.9RR100 pKa = 11.84YY101 pKa = 10.59ASFCFSLYY109 pKa = 9.35QSKK112 pKa = 10.18AASLPAEE119 pKa = 4.2EE120 pKa = 3.94WRR122 pKa = 11.84VRR124 pKa = 11.84SEE126 pKa = 3.62VVAAMRR132 pKa = 11.84RR133 pKa = 11.84LTEE136 pKa = 4.24KK137 pKa = 10.39PDD139 pKa = 3.74PLVPKK144 pKa = 10.15ILTNGARR151 pKa = 11.84LDD153 pKa = 3.76YY154 pKa = 11.0DD155 pKa = 3.89LLEE158 pKa = 4.54EE159 pKa = 4.32AVHH162 pKa = 5.71RR163 pKa = 11.84TIEE166 pKa = 4.42LFSPCNRR173 pKa = 11.84GDD175 pKa = 3.26RR176 pKa = 11.84LARR179 pKa = 11.84RR180 pKa = 11.84RR181 pKa = 11.84LPSRR185 pKa = 11.84GAVLGSSRR193 pKa = 11.84SEE195 pKa = 3.93GGALGQIARR204 pKa = 11.84LGHH207 pKa = 5.88GCDD210 pKa = 3.48FFVPDD215 pKa = 3.98RR216 pKa = 11.84YY217 pKa = 10.86LIGFYY222 pKa = 10.0EE223 pKa = 5.24GPAGAVEE230 pKa = 4.38VRR232 pKa = 11.84SSCEE236 pKa = 3.86PEE238 pKa = 4.03DD239 pKa = 3.77VAWAYY244 pKa = 11.04DD245 pKa = 4.18FFGHH249 pKa = 6.82WGMSHH254 pKa = 7.56DD255 pKa = 4.22VPCWPVGLPEE265 pKa = 4.15PFKK268 pKa = 11.31VRR270 pKa = 11.84VITKK274 pKa = 9.96GYY276 pKa = 9.3APSYY280 pKa = 8.19NHH282 pKa = 6.4ARR284 pKa = 11.84AYY286 pKa = 9.87QPRR289 pKa = 11.84LWRR292 pKa = 11.84SLQEE296 pKa = 4.01HH297 pKa = 5.59NVFRR301 pKa = 11.84LTGEE305 pKa = 4.19MVTGSAIQSFLSEE318 pKa = 4.62CPVDD322 pKa = 3.71SGAFFLSGDD331 pKa = 3.75YY332 pKa = 10.87KK333 pKa = 10.82EE334 pKa = 4.85ATDD337 pKa = 5.24HH338 pKa = 6.59IPSEE342 pKa = 4.4IANLMCMRR350 pKa = 11.84FCDD353 pKa = 3.48RR354 pKa = 11.84LGIPLEE360 pKa = 4.15VVGRR364 pKa = 11.84IARR367 pKa = 11.84ALTGHH372 pKa = 7.43ILPTPEE378 pKa = 5.12LVDD381 pKa = 3.37PTMGLGSQGCMVRR394 pKa = 11.84RR395 pKa = 11.84QKK397 pKa = 10.57KK398 pKa = 7.7GQLMGSPISFPFLCLYY414 pKa = 10.9NLVLQILFHH423 pKa = 6.53FVAEE427 pKa = 4.64GEE429 pKa = 4.35WIRR432 pKa = 11.84DD433 pKa = 3.36LSGVRR438 pKa = 11.84NLINGDD444 pKa = 3.89DD445 pKa = 3.8LLSYY449 pKa = 10.51LRR451 pKa = 11.84EE452 pKa = 4.0RR453 pKa = 11.84GLYY456 pKa = 9.65RR457 pKa = 11.84IWEE460 pKa = 4.28EE461 pKa = 3.87VVEE464 pKa = 4.26WGGLKK469 pKa = 10.39PSVGKK474 pKa = 8.85TIVSQRR480 pKa = 11.84YY481 pKa = 6.82LTINSKK487 pKa = 10.14LFRR490 pKa = 11.84VDD492 pKa = 3.38SAVDD496 pKa = 3.25QHH498 pKa = 5.83GTPYY502 pKa = 9.36WRR504 pKa = 11.84NRR506 pKa = 11.84HH507 pKa = 5.62LPHH510 pKa = 6.3VQLQLAIGSMKK521 pKa = 10.28SGHH524 pKa = 6.48IDD526 pKa = 3.07EE527 pKa = 5.55DD528 pKa = 3.92SWVLSSTSPRR538 pKa = 11.84SRR540 pKa = 11.84SRR542 pKa = 11.84MWQEE546 pKa = 4.41FLDD549 pKa = 4.06SCPDD553 pKa = 3.45KK554 pKa = 11.32DD555 pKa = 4.09RR556 pKa = 11.84AWSFLFSANRR566 pKa = 11.84GLLRR570 pKa = 11.84GLMDD574 pKa = 4.94KK575 pKa = 11.18YY576 pKa = 9.57PTCTLCLPTEE586 pKa = 4.3AGGLGFPLPPIDD598 pKa = 4.2SRR600 pKa = 11.84WYY602 pKa = 7.72VQRR605 pKa = 11.84APRR608 pKa = 11.84SVDD611 pKa = 2.82RR612 pKa = 11.84MKK614 pKa = 11.24ARR616 pKa = 11.84LLLSAGLPSHH626 pKa = 6.96DD627 pKa = 3.72QLRR630 pKa = 11.84SRR632 pKa = 11.84WLSSLASEE640 pKa = 5.43DD641 pKa = 3.84PRR643 pKa = 11.84TADD646 pKa = 3.37ALLFGDD652 pKa = 4.4LLRR655 pKa = 11.84YY656 pKa = 9.03QKK658 pKa = 9.69STGCPLVLEE667 pKa = 4.54TLDD670 pKa = 3.58EE671 pKa = 4.27QVGKK675 pKa = 10.61EE676 pKa = 4.07KK677 pKa = 10.77DD678 pKa = 3.82LPPPILACPLAGGVIDD694 pKa = 4.39PLEE697 pKa = 4.07EE698 pKa = 4.38HH699 pKa = 6.94LLLASQRR706 pKa = 11.84RR707 pKa = 11.84VIRR710 pKa = 11.84DD711 pKa = 2.67IDD713 pKa = 3.65RR714 pKa = 11.84AVRR717 pKa = 11.84AYY719 pKa = 10.35SRR721 pKa = 11.84SGRR724 pKa = 11.84GPWTQCEE731 pKa = 3.75ISDD734 pKa = 3.66FLRR737 pKa = 11.84RR738 pKa = 11.84FAWRR742 pKa = 11.84RR743 pKa = 11.84KK744 pKa = 7.47YY745 pKa = 10.74GRR747 pKa = 11.84YY748 pKa = 9.5LDD750 pKa = 3.93RR751 pKa = 11.84TCGFF755 pKa = 3.9
Molecular weight: 86.0 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
755 |
755 |
755 |
755.0 |
86.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.225 ± 0.0 | 2.384 ± 0.0 |
5.828 ± 0.0 | 5.298 ± 0.0 |
4.503 ± 0.0 | 7.815 ± 0.0 |
2.781 ± 0.0 | 3.709 ± 0.0 |
3.046 ± 0.0 | 12.318 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.987 ± 0.0 | 1.854 ± 0.0 |
6.358 ± 0.0 | 2.914 ± 0.0 |
9.801 ± 0.0 | 7.947 ± 0.0 |
3.576 ± 0.0 | 5.563 ± 0.0 |
2.517 ± 0.0 | 3.576 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
