
Thiocystis violascens (strain ATCC 17096 / DSM 198 / 6111) (Chromatium violascens)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Chromatiales; Chromatiaceae; Thiocystis; Thiocystis violascens
Average proteome isoelectric point is 6.32
Get precalculated fractions of proteins
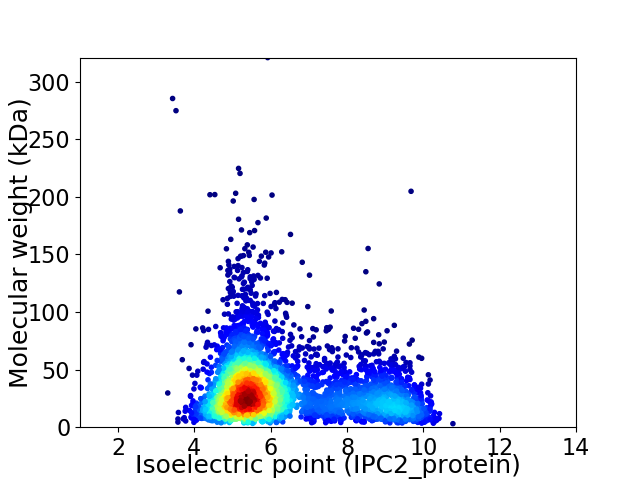
Virtual 2D-PAGE plot for 4192 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
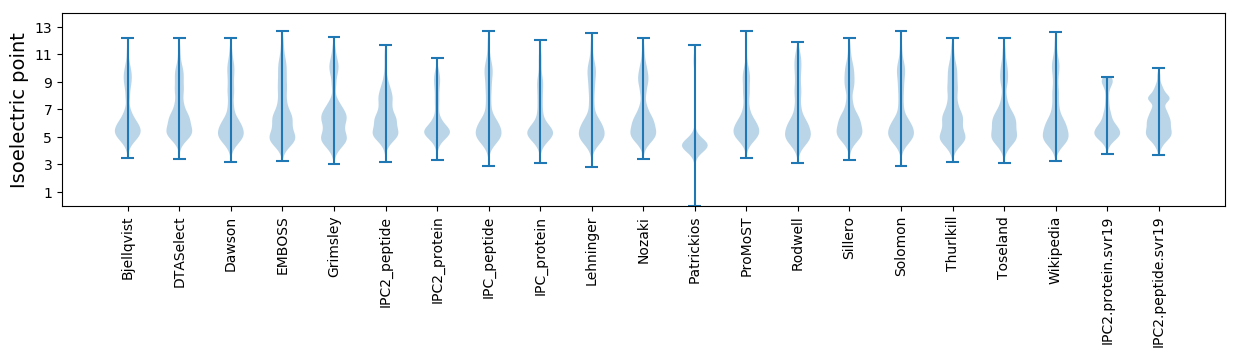
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I3YB38|I3YB38_THIV6 Glycosyl transferase OS=Thiocystis violascens (strain ATCC 17096 / DSM 198 / 6111) OX=765911 GN=Thivi_2257 PE=4 SV=1
MM1 pKa = 7.82DD2 pKa = 5.89RR3 pKa = 11.84SDD5 pKa = 3.77TDD7 pKa = 3.11SDD9 pKa = 3.59TDD11 pKa = 3.54RR12 pKa = 11.84EE13 pKa = 4.28EE14 pKa = 4.38IEE16 pKa = 4.31PVPLATCICQRR27 pKa = 11.84SGFDD31 pKa = 3.04QQARR35 pKa = 11.84LDD37 pKa = 3.97NRR39 pKa = 11.84SSHH42 pKa = 6.95PDD44 pKa = 3.36CSDD47 pKa = 3.23ACCQQGWEE55 pKa = 4.25SSADD59 pKa = 3.72GACQSDD65 pKa = 3.62DD66 pKa = 4.01CEE68 pKa = 5.0FVAQCGGDD76 pKa = 3.21
MM1 pKa = 7.82DD2 pKa = 5.89RR3 pKa = 11.84SDD5 pKa = 3.77TDD7 pKa = 3.11SDD9 pKa = 3.59TDD11 pKa = 3.54RR12 pKa = 11.84EE13 pKa = 4.28EE14 pKa = 4.38IEE16 pKa = 4.31PVPLATCICQRR27 pKa = 11.84SGFDD31 pKa = 3.04QQARR35 pKa = 11.84LDD37 pKa = 3.97NRR39 pKa = 11.84SSHH42 pKa = 6.95PDD44 pKa = 3.36CSDD47 pKa = 3.23ACCQQGWEE55 pKa = 4.25SSADD59 pKa = 3.72GACQSDD65 pKa = 3.62DD66 pKa = 4.01CEE68 pKa = 5.0FVAQCGGDD76 pKa = 3.21
Molecular weight: 8.27 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I3Y5T9|I3Y5T9_THIV6 KaiB domain-containing protein OS=Thiocystis violascens (strain ATCC 17096 / DSM 198 / 6111) OX=765911 GN=Thivi_0288 PE=4 SV=1
MM1 pKa = 7.86DD2 pKa = 3.51VFLIVEE8 pKa = 4.34QILGLVWWLIPLALLVGLLKK28 pKa = 10.83SPSVKK33 pKa = 10.45GYY35 pKa = 10.1LGEE38 pKa = 4.15SRR40 pKa = 11.84VRR42 pKa = 11.84RR43 pKa = 11.84IARR46 pKa = 11.84TRR48 pKa = 11.84LDD50 pKa = 3.18PMTYY54 pKa = 10.16HH55 pKa = 7.42PLHH58 pKa = 6.63NLTLPTPDD66 pKa = 2.96GTTQIDD72 pKa = 3.74HH73 pKa = 6.06VFVSRR78 pKa = 11.84FGLFVLEE85 pKa = 4.47TKK87 pKa = 10.99NMTGWIFGGEE97 pKa = 4.16HH98 pKa = 4.89QAQWTQKK105 pKa = 9.09IYY107 pKa = 10.96RR108 pKa = 11.84RR109 pKa = 11.84TTRR112 pKa = 11.84FQNPLRR118 pKa = 11.84QNYY121 pKa = 8.6KK122 pKa = 9.71HH123 pKa = 6.42LKK125 pKa = 8.26ALEE128 pKa = 4.18RR129 pKa = 11.84ALPVPLEE136 pKa = 4.52TIHH139 pKa = 6.93SVVAFVGDD147 pKa = 3.86STFKK151 pKa = 10.85TEE153 pKa = 3.95MPPNVTQGNGFVRR166 pKa = 11.84HH167 pKa = 5.3IQSFRR172 pKa = 11.84EE173 pKa = 3.94PVFTEE178 pKa = 4.09TQVVEE183 pKa = 3.95LVRR186 pKa = 11.84RR187 pKa = 11.84LQDD190 pKa = 2.96GRR192 pKa = 11.84LAPTLATHH200 pKa = 6.91RR201 pKa = 11.84AHH203 pKa = 6.81VRR205 pKa = 11.84SLKK208 pKa = 10.03QRR210 pKa = 11.84SNPDD214 pKa = 2.69ASQRR218 pKa = 11.84CPRR221 pKa = 11.84CGRR224 pKa = 11.84PMILRR229 pKa = 11.84TVKK232 pKa = 10.39KK233 pKa = 8.49GTQAGRR239 pKa = 11.84RR240 pKa = 11.84FWACSGFPDD249 pKa = 4.29CQSTRR254 pKa = 11.84DD255 pKa = 3.68LPP257 pKa = 4.04
MM1 pKa = 7.86DD2 pKa = 3.51VFLIVEE8 pKa = 4.34QILGLVWWLIPLALLVGLLKK28 pKa = 10.83SPSVKK33 pKa = 10.45GYY35 pKa = 10.1LGEE38 pKa = 4.15SRR40 pKa = 11.84VRR42 pKa = 11.84RR43 pKa = 11.84IARR46 pKa = 11.84TRR48 pKa = 11.84LDD50 pKa = 3.18PMTYY54 pKa = 10.16HH55 pKa = 7.42PLHH58 pKa = 6.63NLTLPTPDD66 pKa = 2.96GTTQIDD72 pKa = 3.74HH73 pKa = 6.06VFVSRR78 pKa = 11.84FGLFVLEE85 pKa = 4.47TKK87 pKa = 10.99NMTGWIFGGEE97 pKa = 4.16HH98 pKa = 4.89QAQWTQKK105 pKa = 9.09IYY107 pKa = 10.96RR108 pKa = 11.84RR109 pKa = 11.84TTRR112 pKa = 11.84FQNPLRR118 pKa = 11.84QNYY121 pKa = 8.6KK122 pKa = 9.71HH123 pKa = 6.42LKK125 pKa = 8.26ALEE128 pKa = 4.18RR129 pKa = 11.84ALPVPLEE136 pKa = 4.52TIHH139 pKa = 6.93SVVAFVGDD147 pKa = 3.86STFKK151 pKa = 10.85TEE153 pKa = 3.95MPPNVTQGNGFVRR166 pKa = 11.84HH167 pKa = 5.3IQSFRR172 pKa = 11.84EE173 pKa = 3.94PVFTEE178 pKa = 4.09TQVVEE183 pKa = 3.95LVRR186 pKa = 11.84RR187 pKa = 11.84LQDD190 pKa = 2.96GRR192 pKa = 11.84LAPTLATHH200 pKa = 6.91RR201 pKa = 11.84AHH203 pKa = 6.81VRR205 pKa = 11.84SLKK208 pKa = 10.03QRR210 pKa = 11.84SNPDD214 pKa = 2.69ASQRR218 pKa = 11.84CPRR221 pKa = 11.84CGRR224 pKa = 11.84PMILRR229 pKa = 11.84TVKK232 pKa = 10.39KK233 pKa = 8.49GTQAGRR239 pKa = 11.84RR240 pKa = 11.84FWACSGFPDD249 pKa = 4.29CQSTRR254 pKa = 11.84DD255 pKa = 3.68LPP257 pKa = 4.04
Molecular weight: 29.43 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1375469 |
26 |
2896 |
328.1 |
36.05 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.428 ± 0.047 | 1.003 ± 0.013 |
6.006 ± 0.031 | 6.148 ± 0.037 |
3.486 ± 0.02 | 7.925 ± 0.035 |
2.229 ± 0.019 | 4.987 ± 0.028 |
2.733 ± 0.029 | 11.489 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.191 ± 0.018 | 2.553 ± 0.023 |
5.312 ± 0.032 | 3.717 ± 0.027 |
7.969 ± 0.044 | 5.242 ± 0.027 |
5.117 ± 0.033 | 6.77 ± 0.033 |
1.404 ± 0.017 | 2.289 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
