
Methanobrevibacter filiformis
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Methanomada group; Methanobacteria; Methanobacteriales; Methanobacteriaceae; Methanobrevibacter
Average proteome isoelectric point is 6.36
Get precalculated fractions of proteins
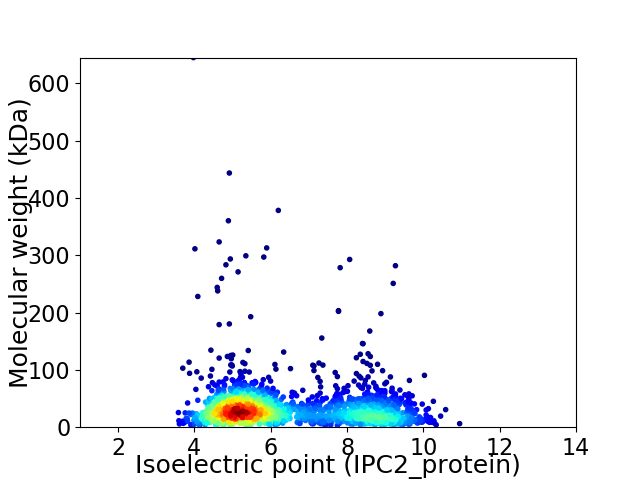
Virtual 2D-PAGE plot for 1920 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A166FFZ0|A0A166FFZ0_9EURY Uncharacterized protein OS=Methanobrevibacter filiformis OX=55758 GN=MBFIL_00230 PE=4 SV=1
MM1 pKa = 8.21AEE3 pKa = 3.81EE4 pKa = 5.8DD5 pKa = 3.82YY6 pKa = 11.6DD7 pKa = 4.34FDD9 pKa = 5.36EE10 pKa = 4.57EE11 pKa = 4.8TEE13 pKa = 4.56DD14 pKa = 3.59SLKK17 pKa = 10.38MFTGSVIDD25 pKa = 3.38IQYY28 pKa = 10.97LVDD31 pKa = 5.75NMFLDD36 pKa = 4.11YY37 pKa = 10.59KK38 pKa = 11.15SKK40 pKa = 11.07FEE42 pKa = 4.07NQEE45 pKa = 3.96SPISYY50 pKa = 10.06LDD52 pKa = 3.35EE53 pKa = 6.28GGDD56 pKa = 3.52ARR58 pKa = 11.84NLFEE62 pKa = 5.08TISLPLFEE70 pKa = 5.35IFWLINEE77 pKa = 4.44SVLMKK82 pKa = 9.06YY83 pKa = 9.89IPYY86 pKa = 10.52ASGDD90 pKa = 3.57FLDD93 pKa = 5.06ILGHH97 pKa = 5.78NNPRR101 pKa = 11.84NPGKK105 pKa = 10.38NSTGQLQFSLPHH117 pKa = 6.96DD118 pKa = 3.65VLKK121 pKa = 10.84DD122 pKa = 3.53YY123 pKa = 10.98DD124 pKa = 3.84ITIPAYY130 pKa = 10.07SVYY133 pKa = 9.82LTEE136 pKa = 6.15DD137 pKa = 3.64DD138 pKa = 5.49LGLEE142 pKa = 4.27YY143 pKa = 9.82EE144 pKa = 4.64TIEE147 pKa = 4.64EE148 pKa = 4.8AILPAGEE155 pKa = 4.05NSILVPARR163 pKa = 11.84SVFGGSEE170 pKa = 4.03YY171 pKa = 11.01NIDD174 pKa = 3.91SNLVTIFEE182 pKa = 4.19EE183 pKa = 4.93SIDD186 pKa = 3.98DD187 pKa = 4.26LLVTNPVPFIGGTDD201 pKa = 3.76DD202 pKa = 5.64EE203 pKa = 5.78DD204 pKa = 4.71DD205 pKa = 3.62EE206 pKa = 5.11YY207 pKa = 11.93YY208 pKa = 10.38RR209 pKa = 11.84ARR211 pKa = 11.84LLYY214 pKa = 9.43EE215 pKa = 3.78QSEE218 pKa = 4.46HH219 pKa = 7.55DD220 pKa = 3.82FGSVGWYY227 pKa = 9.92KK228 pKa = 10.79RR229 pKa = 11.84IAVKK233 pKa = 10.39IPGVHH238 pKa = 6.29DD239 pKa = 3.69VKK241 pKa = 11.24VINCLKK247 pKa = 10.23GDD249 pKa = 3.73YY250 pKa = 10.14TIGIIVNPPTAEE262 pKa = 3.38IVQNVTNFFNTADD275 pKa = 3.72TTPAGINAYY284 pKa = 9.38IYY286 pKa = 10.38GAEE289 pKa = 4.19TFPVDD294 pKa = 5.5IIIQDD299 pKa = 3.76LVFTNEE305 pKa = 3.69VNPADD310 pKa = 3.73VVEE313 pKa = 5.21EE314 pKa = 4.01IQNQINKK321 pKa = 9.33YY322 pKa = 9.77FNNLNISEE330 pKa = 4.49TIYY333 pKa = 10.83RR334 pKa = 11.84NSILSILSNINGLIDD349 pKa = 3.56YY350 pKa = 7.46TLISPSVNIDD360 pKa = 3.22AEE362 pKa = 4.44EE363 pKa = 4.1NQVLIQGEE371 pKa = 4.45TIINN375 pKa = 3.56
MM1 pKa = 8.21AEE3 pKa = 3.81EE4 pKa = 5.8DD5 pKa = 3.82YY6 pKa = 11.6DD7 pKa = 4.34FDD9 pKa = 5.36EE10 pKa = 4.57EE11 pKa = 4.8TEE13 pKa = 4.56DD14 pKa = 3.59SLKK17 pKa = 10.38MFTGSVIDD25 pKa = 3.38IQYY28 pKa = 10.97LVDD31 pKa = 5.75NMFLDD36 pKa = 4.11YY37 pKa = 10.59KK38 pKa = 11.15SKK40 pKa = 11.07FEE42 pKa = 4.07NQEE45 pKa = 3.96SPISYY50 pKa = 10.06LDD52 pKa = 3.35EE53 pKa = 6.28GGDD56 pKa = 3.52ARR58 pKa = 11.84NLFEE62 pKa = 5.08TISLPLFEE70 pKa = 5.35IFWLINEE77 pKa = 4.44SVLMKK82 pKa = 9.06YY83 pKa = 9.89IPYY86 pKa = 10.52ASGDD90 pKa = 3.57FLDD93 pKa = 5.06ILGHH97 pKa = 5.78NNPRR101 pKa = 11.84NPGKK105 pKa = 10.38NSTGQLQFSLPHH117 pKa = 6.96DD118 pKa = 3.65VLKK121 pKa = 10.84DD122 pKa = 3.53YY123 pKa = 10.98DD124 pKa = 3.84ITIPAYY130 pKa = 10.07SVYY133 pKa = 9.82LTEE136 pKa = 6.15DD137 pKa = 3.64DD138 pKa = 5.49LGLEE142 pKa = 4.27YY143 pKa = 9.82EE144 pKa = 4.64TIEE147 pKa = 4.64EE148 pKa = 4.8AILPAGEE155 pKa = 4.05NSILVPARR163 pKa = 11.84SVFGGSEE170 pKa = 4.03YY171 pKa = 11.01NIDD174 pKa = 3.91SNLVTIFEE182 pKa = 4.19EE183 pKa = 4.93SIDD186 pKa = 3.98DD187 pKa = 4.26LLVTNPVPFIGGTDD201 pKa = 3.76DD202 pKa = 5.64EE203 pKa = 5.78DD204 pKa = 4.71DD205 pKa = 3.62EE206 pKa = 5.11YY207 pKa = 11.93YY208 pKa = 10.38RR209 pKa = 11.84ARR211 pKa = 11.84LLYY214 pKa = 9.43EE215 pKa = 3.78QSEE218 pKa = 4.46HH219 pKa = 7.55DD220 pKa = 3.82FGSVGWYY227 pKa = 9.92KK228 pKa = 10.79RR229 pKa = 11.84IAVKK233 pKa = 10.39IPGVHH238 pKa = 6.29DD239 pKa = 3.69VKK241 pKa = 11.24VINCLKK247 pKa = 10.23GDD249 pKa = 3.73YY250 pKa = 10.14TIGIIVNPPTAEE262 pKa = 3.38IVQNVTNFFNTADD275 pKa = 3.72TTPAGINAYY284 pKa = 9.38IYY286 pKa = 10.38GAEE289 pKa = 4.19TFPVDD294 pKa = 5.5IIIQDD299 pKa = 3.76LVFTNEE305 pKa = 3.69VNPADD310 pKa = 3.73VVEE313 pKa = 5.21EE314 pKa = 4.01IQNQINKK321 pKa = 9.33YY322 pKa = 9.77FNNLNISEE330 pKa = 4.49TIYY333 pKa = 10.83RR334 pKa = 11.84NSILSILSNINGLIDD349 pKa = 3.56YY350 pKa = 7.46TLISPSVNIDD360 pKa = 3.22AEE362 pKa = 4.44EE363 pKa = 4.1NQVLIQGEE371 pKa = 4.45TIINN375 pKa = 3.56
Molecular weight: 42.24 kDa
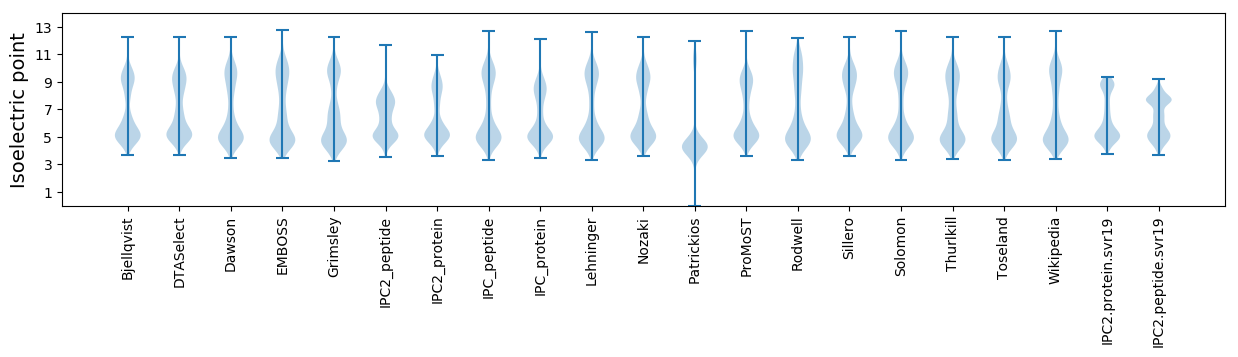
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A166ESV9|A0A166ESV9_9EURY Modification methylase HaeIII OS=Methanobrevibacter filiformis OX=55758 GN=haeIIIM PE=3 SV=1
MM1 pKa = 6.68VRR3 pKa = 11.84AYY5 pKa = 10.52SRR7 pKa = 11.84RR8 pKa = 11.84EE9 pKa = 3.95YY10 pKa = 10.02IRR12 pKa = 11.84KK13 pKa = 9.13IPGSRR18 pKa = 11.84IVQYY22 pKa = 11.77DD23 pKa = 3.51MGNLSQRR30 pKa = 11.84FPVAVSLAVKK40 pKa = 8.41TPARR44 pKa = 11.84ITHH47 pKa = 6.32NALEE51 pKa = 4.49AGRR54 pKa = 11.84VASNRR59 pKa = 11.84FMQRR63 pKa = 11.84KK64 pKa = 8.84AGRR67 pKa = 11.84LGYY70 pKa = 9.36HH71 pKa = 6.23LKK73 pKa = 10.25IRR75 pKa = 11.84VYY77 pKa = 7.96PHH79 pKa = 6.37NIIRR83 pKa = 11.84EE84 pKa = 4.11NPMATGAGADD94 pKa = 3.36RR95 pKa = 11.84VQDD98 pKa = 3.52GMRR101 pKa = 11.84KK102 pKa = 9.49AFGKK106 pKa = 10.01SVSSEE111 pKa = 3.7AVVKK115 pKa = 10.35RR116 pKa = 11.84NQRR119 pKa = 11.84IFSIYY124 pKa = 9.11TSEE127 pKa = 4.98KK128 pKa = 9.8NFGDD132 pKa = 3.26AKK134 pKa = 10.27EE135 pKa = 4.24SLRR138 pKa = 11.84RR139 pKa = 11.84AAMKK143 pKa = 10.08MPVSCKK149 pKa = 9.96IVIDD153 pKa = 3.99EE154 pKa = 4.21GAEE157 pKa = 3.98LVKK160 pKa = 11.05
MM1 pKa = 6.68VRR3 pKa = 11.84AYY5 pKa = 10.52SRR7 pKa = 11.84RR8 pKa = 11.84EE9 pKa = 3.95YY10 pKa = 10.02IRR12 pKa = 11.84KK13 pKa = 9.13IPGSRR18 pKa = 11.84IVQYY22 pKa = 11.77DD23 pKa = 3.51MGNLSQRR30 pKa = 11.84FPVAVSLAVKK40 pKa = 8.41TPARR44 pKa = 11.84ITHH47 pKa = 6.32NALEE51 pKa = 4.49AGRR54 pKa = 11.84VASNRR59 pKa = 11.84FMQRR63 pKa = 11.84KK64 pKa = 8.84AGRR67 pKa = 11.84LGYY70 pKa = 9.36HH71 pKa = 6.23LKK73 pKa = 10.25IRR75 pKa = 11.84VYY77 pKa = 7.96PHH79 pKa = 6.37NIIRR83 pKa = 11.84EE84 pKa = 4.11NPMATGAGADD94 pKa = 3.36RR95 pKa = 11.84VQDD98 pKa = 3.52GMRR101 pKa = 11.84KK102 pKa = 9.49AFGKK106 pKa = 10.01SVSSEE111 pKa = 3.7AVVKK115 pKa = 10.35RR116 pKa = 11.84NQRR119 pKa = 11.84IFSIYY124 pKa = 9.11TSEE127 pKa = 4.98KK128 pKa = 9.8NFGDD132 pKa = 3.26AKK134 pKa = 10.27EE135 pKa = 4.24SLRR138 pKa = 11.84RR139 pKa = 11.84AAMKK143 pKa = 10.08MPVSCKK149 pKa = 9.96IVIDD153 pKa = 3.99EE154 pKa = 4.21GAEE157 pKa = 3.98LVKK160 pKa = 11.05
Molecular weight: 18.01 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
604930 |
29 |
6034 |
315.1 |
35.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.298 ± 0.061 | 0.909 ± 0.027 |
5.617 ± 0.064 | 6.491 ± 0.133 |
4.211 ± 0.041 | 6.573 ± 0.1 |
1.523 ± 0.028 | 10.254 ± 0.056 |
8.491 ± 0.156 | 8.373 ± 0.085 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.93 ± 0.036 | 8.722 ± 0.25 |
2.864 ± 0.05 | 1.989 ± 0.04 |
2.776 ± 0.058 | 7.21 ± 0.094 |
5.955 ± 0.14 | 6.309 ± 0.084 |
0.656 ± 0.016 | 3.848 ± 0.043 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
