
Capybara microvirus Cap3_SP_442
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.54
Get precalculated fractions of proteins
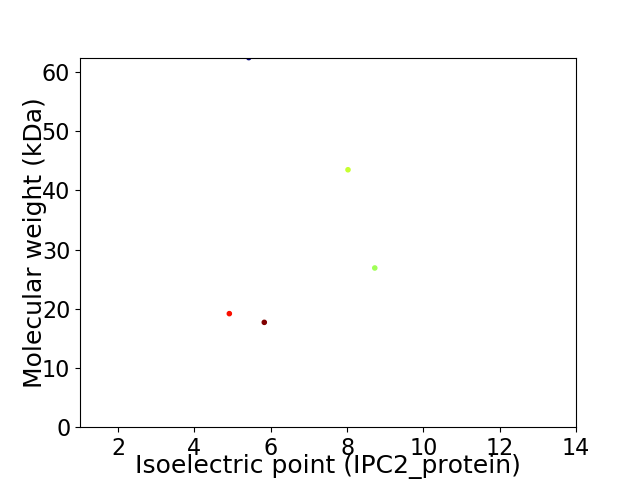
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
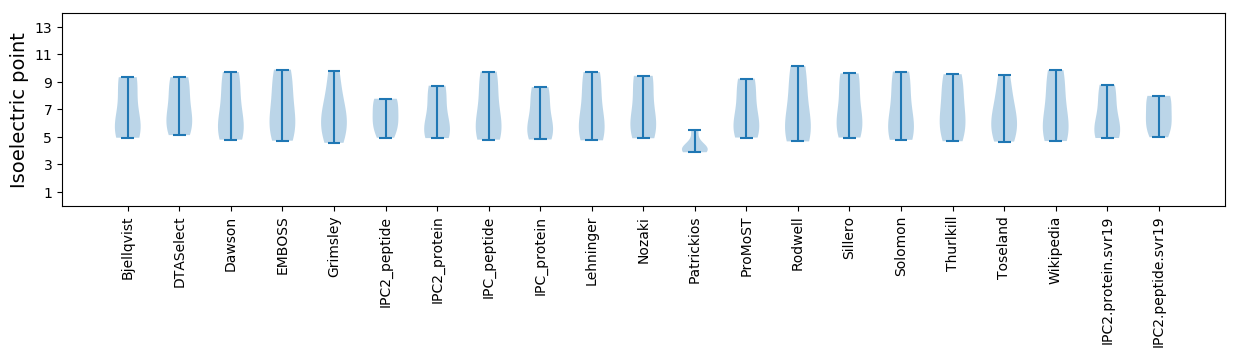
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W550|A0A4P8W550_9VIRU Major capsid protein OS=Capybara microvirus Cap3_SP_442 OX=2585454 PE=3 SV=1
MM1 pKa = 7.75RR2 pKa = 11.84DD3 pKa = 3.45VVKK6 pKa = 10.78KK7 pKa = 10.0MCDD10 pKa = 2.95EE11 pKa = 4.17LTAHH15 pKa = 6.39GVVFSLSEE23 pKa = 4.19VKK25 pKa = 10.56KK26 pKa = 10.04LTDD29 pKa = 3.37AQVNCVLEE37 pKa = 4.59MIASCKK43 pKa = 9.27EE44 pKa = 3.69PSVIASFIATASRR57 pKa = 11.84CSVNISSEE65 pKa = 4.14DD66 pKa = 3.49VAKK69 pKa = 11.03QNDD72 pKa = 3.56DD73 pKa = 3.29HH74 pKa = 7.56QVYY77 pKa = 10.45VDD79 pKa = 3.68EE80 pKa = 6.17DD81 pKa = 3.88KK82 pKa = 11.4LKK84 pKa = 11.05SDD86 pKa = 3.93ANVVGLFGVFDD97 pKa = 3.88RR98 pKa = 11.84QSNFIRR104 pKa = 11.84SWFFCDD110 pKa = 4.02NEE112 pKa = 4.14VQARR116 pKa = 11.84RR117 pKa = 11.84FFKK120 pKa = 10.94LSASKK125 pKa = 10.84LDD127 pKa = 3.54ADD129 pKa = 3.92IVLSPEE135 pKa = 4.48DD136 pKa = 3.72FDD138 pKa = 5.74LVLLAALGRR147 pKa = 11.84SGLVQPLASRR157 pKa = 11.84VVFNFAEE164 pKa = 4.36LFDD167 pKa = 4.19KK168 pKa = 11.09QLDD171 pKa = 3.86KK172 pKa = 11.65
MM1 pKa = 7.75RR2 pKa = 11.84DD3 pKa = 3.45VVKK6 pKa = 10.78KK7 pKa = 10.0MCDD10 pKa = 2.95EE11 pKa = 4.17LTAHH15 pKa = 6.39GVVFSLSEE23 pKa = 4.19VKK25 pKa = 10.56KK26 pKa = 10.04LTDD29 pKa = 3.37AQVNCVLEE37 pKa = 4.59MIASCKK43 pKa = 9.27EE44 pKa = 3.69PSVIASFIATASRR57 pKa = 11.84CSVNISSEE65 pKa = 4.14DD66 pKa = 3.49VAKK69 pKa = 11.03QNDD72 pKa = 3.56DD73 pKa = 3.29HH74 pKa = 7.56QVYY77 pKa = 10.45VDD79 pKa = 3.68EE80 pKa = 6.17DD81 pKa = 3.88KK82 pKa = 11.4LKK84 pKa = 11.05SDD86 pKa = 3.93ANVVGLFGVFDD97 pKa = 3.88RR98 pKa = 11.84QSNFIRR104 pKa = 11.84SWFFCDD110 pKa = 4.02NEE112 pKa = 4.14VQARR116 pKa = 11.84RR117 pKa = 11.84FFKK120 pKa = 10.94LSASKK125 pKa = 10.84LDD127 pKa = 3.54ADD129 pKa = 3.92IVLSPEE135 pKa = 4.48DD136 pKa = 3.72FDD138 pKa = 5.74LVLLAALGRR147 pKa = 11.84SGLVQPLASRR157 pKa = 11.84VVFNFAEE164 pKa = 4.36LFDD167 pKa = 4.19KK168 pKa = 11.09QLDD171 pKa = 3.86KK172 pKa = 11.65
Molecular weight: 19.18 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W4U1|A0A4P8W4U1_9VIRU Minor capsid protein OS=Capybara microvirus Cap3_SP_442 OX=2585454 PE=4 SV=1
MM1 pKa = 7.41SLLAAAAGNIFDD13 pKa = 3.98RR14 pKa = 11.84ALNIGEE20 pKa = 4.33GAATSAINSAIGWKK34 pKa = 7.83YY35 pKa = 9.34QKK37 pKa = 11.02KK38 pKa = 9.98AMNYY42 pKa = 9.08SDD44 pKa = 3.29QLQRR48 pKa = 11.84SFAHH52 pKa = 6.83DD53 pKa = 3.12SYY55 pKa = 11.83GIMRR59 pKa = 11.84SGLEE63 pKa = 3.85DD64 pKa = 3.16AGYY67 pKa = 11.0NPLLSLSGSSNSEE80 pKa = 3.8PLVAGASSASGTGSGQGMGNAAAMKK105 pKa = 8.83QAKK108 pKa = 9.39AAEE111 pKa = 4.22EE112 pKa = 4.23LKK114 pKa = 10.91DD115 pKa = 3.87SQIDD119 pKa = 3.75LNASQEE125 pKa = 4.21ALNGTNSAKK134 pKa = 10.57AQADD138 pKa = 3.88INNSTAMTKK147 pKa = 10.5AQVDD151 pKa = 3.97LAGSQSLLNGMNSAKK166 pKa = 10.55SQVDD170 pKa = 3.65MKK172 pKa = 11.25KK173 pKa = 10.56LLADD177 pKa = 3.92IKK179 pKa = 11.17NNTAITKK186 pKa = 9.81AQVGLINAQAKK197 pKa = 8.14AARR200 pKa = 11.84YY201 pKa = 8.49GLAGNEE207 pKa = 3.79YY208 pKa = 10.26EE209 pKa = 4.35AMKK212 pKa = 10.81NKK214 pKa = 10.3LYY216 pKa = 9.93STLAKK221 pKa = 10.19EE222 pKa = 4.28NPNAFKK228 pKa = 11.17AMLVGQGATSTAKK241 pKa = 10.94GIGEE245 pKa = 4.32ALRR248 pKa = 11.84SFLPKK253 pKa = 10.37FSVSFGKK260 pKa = 10.44
MM1 pKa = 7.41SLLAAAAGNIFDD13 pKa = 3.98RR14 pKa = 11.84ALNIGEE20 pKa = 4.33GAATSAINSAIGWKK34 pKa = 7.83YY35 pKa = 9.34QKK37 pKa = 11.02KK38 pKa = 9.98AMNYY42 pKa = 9.08SDD44 pKa = 3.29QLQRR48 pKa = 11.84SFAHH52 pKa = 6.83DD53 pKa = 3.12SYY55 pKa = 11.83GIMRR59 pKa = 11.84SGLEE63 pKa = 3.85DD64 pKa = 3.16AGYY67 pKa = 11.0NPLLSLSGSSNSEE80 pKa = 3.8PLVAGASSASGTGSGQGMGNAAAMKK105 pKa = 8.83QAKK108 pKa = 9.39AAEE111 pKa = 4.22EE112 pKa = 4.23LKK114 pKa = 10.91DD115 pKa = 3.87SQIDD119 pKa = 3.75LNASQEE125 pKa = 4.21ALNGTNSAKK134 pKa = 10.57AQADD138 pKa = 3.88INNSTAMTKK147 pKa = 10.5AQVDD151 pKa = 3.97LAGSQSLLNGMNSAKK166 pKa = 10.55SQVDD170 pKa = 3.65MKK172 pKa = 11.25KK173 pKa = 10.56LLADD177 pKa = 3.92IKK179 pKa = 11.17NNTAITKK186 pKa = 9.81AQVGLINAQAKK197 pKa = 8.14AARR200 pKa = 11.84YY201 pKa = 8.49GLAGNEE207 pKa = 3.79YY208 pKa = 10.26EE209 pKa = 4.35AMKK212 pKa = 10.81NKK214 pKa = 10.3LYY216 pKa = 9.93STLAKK221 pKa = 10.19EE222 pKa = 4.28NPNAFKK228 pKa = 11.17AMLVGQGATSTAKK241 pKa = 10.94GIGEE245 pKa = 4.32ALRR248 pKa = 11.84SFLPKK253 pKa = 10.37FSVSFGKK260 pKa = 10.44
Molecular weight: 26.89 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1509 |
153 |
557 |
301.8 |
33.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.687 ± 2.169 | 1.657 ± 0.454 |
6.428 ± 0.817 | 4.771 ± 0.716 |
5.633 ± 0.683 | 5.964 ± 1.111 |
1.657 ± 0.359 | 4.307 ± 0.397 |
5.898 ± 1.037 | 9.079 ± 0.399 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.651 ± 0.355 | 6.097 ± 0.67 |
4.307 ± 1.105 | 4.838 ± 0.594 |
5.235 ± 0.809 | 9.079 ± 0.565 |
3.91 ± 0.54 | 5.765 ± 1.087 |
0.795 ± 0.205 | 4.241 ± 0.822 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
