
Lucilia cuprina (Green bottle fly) (Australian sheep blowfly)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Ecdysozoa; Panarthropoda; Arthropoda; Mandibulata; Pancrustacea; Hexapoda; Insecta; Dicondylia; Pterygota; Neoptera; Endopterygota; Diptera; Brachycera; Muscomorpha; Eremoneura; Cyclorrhapha;
Average proteome isoelectric point is 6.79
Get precalculated fractions of proteins
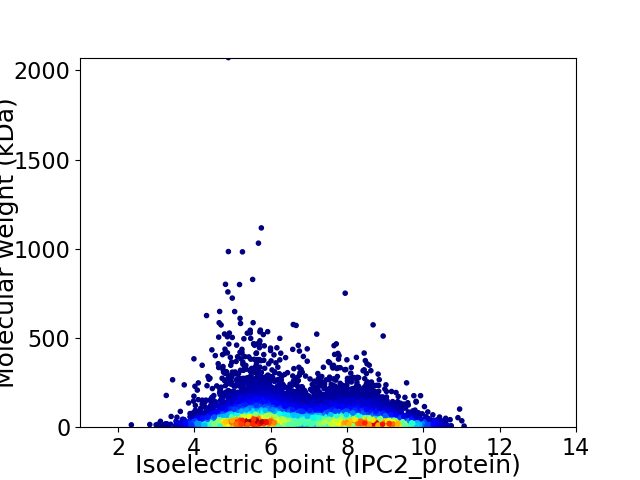
Virtual 2D-PAGE plot for 14353 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0L0BL23|A0A0L0BL23_LUCCU CULLIN_2 domain-containing protein OS=Lucilia cuprina OX=7375 GN=FF38_07904 PE=3 SV=1
MM1 pKa = 7.73SDD3 pKa = 3.23VDD5 pKa = 5.09DD6 pKa = 5.52DD7 pKa = 5.77FNFLNCQWDD16 pKa = 3.72VMIITSGAEE25 pKa = 3.62FLMSINTFDD34 pKa = 4.53SSTLVCLPLPPEE46 pKa = 4.45SPVLACSLCRR56 pKa = 11.84PLEE59 pKa = 4.12PVEE62 pKa = 4.17IAVTTAVPGAGICVTLGGICDD83 pKa = 4.16DD84 pKa = 4.66DD85 pKa = 4.95AVVACVDD92 pKa = 3.65DD93 pKa = 4.2AVVVVVDD100 pKa = 4.2DD101 pKa = 4.54GVAVFAFVVAVVNVADD117 pKa = 5.56AGDD120 pKa = 4.25DD121 pKa = 3.56DD122 pKa = 5.65DD123 pKa = 5.14RR124 pKa = 11.84FPVRR128 pKa = 11.84VYY130 pKa = 10.95DD131 pKa = 3.78VNADD135 pKa = 3.56VAVDD139 pKa = 3.52VAVDD143 pKa = 3.64VVVDD147 pKa = 3.66VALASSSLDD156 pKa = 3.23SCGRR160 pKa = 11.84CILRR164 pKa = 11.84LDD166 pKa = 3.85NVDD169 pKa = 3.47TVGGGGGAICAASYY183 pKa = 10.24PKK185 pKa = 10.32VEE187 pKa = 4.09KK188 pKa = 10.55RR189 pKa = 11.84GGGGDD194 pKa = 3.92TIDD197 pKa = 3.21MLSRR201 pKa = 11.84EE202 pKa = 4.26RR203 pKa = 11.84AFEE206 pKa = 3.88LRR208 pKa = 11.84PISSNLDD215 pKa = 3.48SLDD218 pKa = 3.2WQYY221 pKa = 12.0VKK223 pKa = 10.84LVAGLGNSFEE233 pKa = 4.49LYY235 pKa = 10.18LYY237 pKa = 10.85NFLEE241 pKa = 4.75SKK243 pKa = 10.29PQDD246 pKa = 3.24TSAHH250 pKa = 5.87HH251 pKa = 6.7LCFFF255 pKa = 5.08
MM1 pKa = 7.73SDD3 pKa = 3.23VDD5 pKa = 5.09DD6 pKa = 5.52DD7 pKa = 5.77FNFLNCQWDD16 pKa = 3.72VMIITSGAEE25 pKa = 3.62FLMSINTFDD34 pKa = 4.53SSTLVCLPLPPEE46 pKa = 4.45SPVLACSLCRR56 pKa = 11.84PLEE59 pKa = 4.12PVEE62 pKa = 4.17IAVTTAVPGAGICVTLGGICDD83 pKa = 4.16DD84 pKa = 4.66DD85 pKa = 4.95AVVACVDD92 pKa = 3.65DD93 pKa = 4.2AVVVVVDD100 pKa = 4.2DD101 pKa = 4.54GVAVFAFVVAVVNVADD117 pKa = 5.56AGDD120 pKa = 4.25DD121 pKa = 3.56DD122 pKa = 5.65DD123 pKa = 5.14RR124 pKa = 11.84FPVRR128 pKa = 11.84VYY130 pKa = 10.95DD131 pKa = 3.78VNADD135 pKa = 3.56VAVDD139 pKa = 3.52VAVDD143 pKa = 3.64VVVDD147 pKa = 3.66VALASSSLDD156 pKa = 3.23SCGRR160 pKa = 11.84CILRR164 pKa = 11.84LDD166 pKa = 3.85NVDD169 pKa = 3.47TVGGGGGAICAASYY183 pKa = 10.24PKK185 pKa = 10.32VEE187 pKa = 4.09KK188 pKa = 10.55RR189 pKa = 11.84GGGGDD194 pKa = 3.92TIDD197 pKa = 3.21MLSRR201 pKa = 11.84EE202 pKa = 4.26RR203 pKa = 11.84AFEE206 pKa = 3.88LRR208 pKa = 11.84PISSNLDD215 pKa = 3.48SLDD218 pKa = 3.2WQYY221 pKa = 12.0VKK223 pKa = 10.84LVAGLGNSFEE233 pKa = 4.49LYY235 pKa = 10.18LYY237 pKa = 10.85NFLEE241 pKa = 4.75SKK243 pKa = 10.29PQDD246 pKa = 3.24TSAHH250 pKa = 5.87HH251 pKa = 6.7LCFFF255 pKa = 5.08
Molecular weight: 26.99 kDa
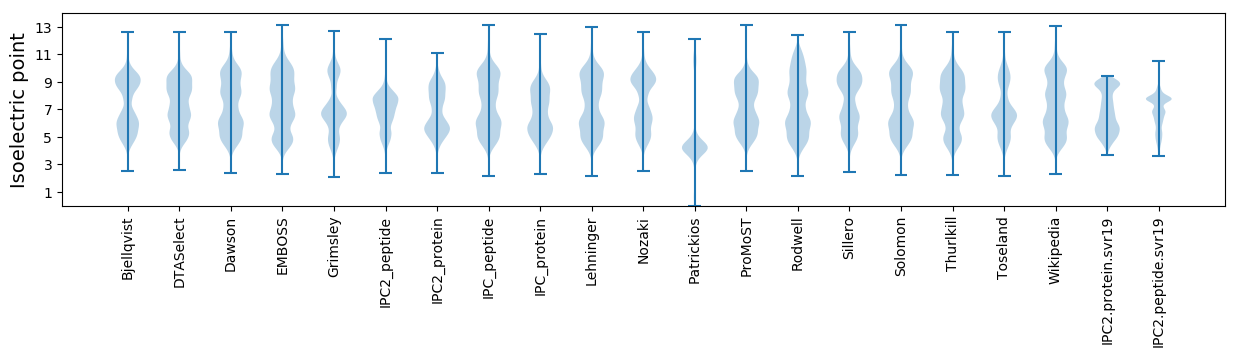
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0L0CSZ1|A0A0L0CSZ1_LUCCU SUEL-type lectin domain-containing protein (Fragment) OS=Lucilia cuprina OX=7375 GN=FF38_02861 PE=4 SV=1
MM1 pKa = 7.21MSSTLSKK8 pKa = 10.35IHH10 pKa = 5.38VTNSTGLSLNEE21 pKa = 3.83RR22 pKa = 11.84FTAISTQPKK31 pKa = 8.77VVTNIGATRR40 pKa = 11.84RR41 pKa = 11.84SRR43 pKa = 11.84SRR45 pKa = 11.84SRR47 pKa = 11.84SVQRR51 pKa = 11.84QVPVAAAAEE60 pKa = 4.73RR61 pKa = 11.84ITSRR65 pKa = 11.84ANQRR69 pKa = 11.84LLAQLQRR76 pKa = 11.84KK77 pKa = 8.82HH78 pKa = 6.1KK79 pKa = 9.9VQAALKK85 pKa = 9.19LKK87 pKa = 9.02RR88 pKa = 11.84RR89 pKa = 11.84SMRR92 pKa = 11.84TVGGRR97 pKa = 11.84SNGSGGANGRR107 pKa = 11.84IRR109 pKa = 11.84RR110 pKa = 11.84GTVKK114 pKa = 10.58AFRR117 pKa = 11.84VGANGKK123 pKa = 8.74PIRR126 pKa = 11.84TNSLTRR132 pKa = 11.84IATMNADD139 pKa = 4.4RR140 pKa = 11.84EE141 pKa = 4.48ITNNRR146 pKa = 11.84SRR148 pKa = 11.84PLRR151 pKa = 11.84RR152 pKa = 11.84SNSSNNLAGRR162 pKa = 11.84LGPRR166 pKa = 11.84RR167 pKa = 11.84PTVGGAAQRR176 pKa = 11.84VARR179 pKa = 11.84RR180 pKa = 11.84NIDD183 pKa = 3.38RR184 pKa = 11.84QRR186 pKa = 11.84GRR188 pKa = 11.84NNVGIRR194 pKa = 11.84GRR196 pKa = 11.84STGINNRR203 pKa = 11.84GRR205 pKa = 11.84SRR207 pKa = 11.84SRR209 pKa = 11.84SRR211 pKa = 11.84TRR213 pKa = 11.84TLSDD217 pKa = 2.75NGRR220 pKa = 11.84NRR222 pKa = 11.84SRR224 pKa = 11.84TRR226 pKa = 11.84NNTDD230 pKa = 2.65NQLNNRR236 pKa = 11.84SRR238 pKa = 11.84SRR240 pKa = 11.84NRR242 pKa = 11.84NSANIQTNNNNNNVRR257 pKa = 11.84RR258 pKa = 11.84GRR260 pKa = 11.84SRR262 pKa = 11.84SRR264 pKa = 11.84SRR266 pKa = 11.84GRR268 pKa = 11.84AAVGSVNKK276 pKa = 10.54SNLPIKK282 pKa = 10.41ARR284 pKa = 11.84LGVRR288 pKa = 11.84PGVTANRR295 pKa = 11.84GTNNNRR301 pKa = 11.84RR302 pKa = 11.84GRR304 pKa = 11.84NASLTRR310 pKa = 11.84WGVQTGRR317 pKa = 11.84VQKK320 pKa = 10.39RR321 pKa = 11.84RR322 pKa = 11.84NSTNVNAAGNDD333 pKa = 3.42NNGRR337 pKa = 11.84GRR339 pKa = 11.84MRR341 pKa = 11.84SRR343 pKa = 11.84SAVRR347 pKa = 11.84NNSNNGNVRR356 pKa = 11.84SRR358 pKa = 11.84SRR360 pKa = 11.84SRR362 pKa = 11.84TRR364 pKa = 11.84KK365 pKa = 9.23GQVGQQRR372 pKa = 11.84QQQGSGQVRR381 pKa = 11.84QQRR384 pKa = 11.84NNRR387 pKa = 11.84APRR390 pKa = 11.84GRR392 pKa = 11.84QQQRR396 pKa = 11.84RR397 pKa = 11.84GNNTGNGQRR406 pKa = 11.84GTNNNGKK413 pKa = 9.07AQTQQRR419 pKa = 11.84RR420 pKa = 11.84GRR422 pKa = 11.84SRR424 pKa = 11.84SRR426 pKa = 11.84TGRR429 pKa = 11.84GANSGGNSRR438 pKa = 11.84NQGNKK443 pKa = 9.04SAAATKK449 pKa = 9.88PAVNKK454 pKa = 10.21DD455 pKa = 3.63ALDD458 pKa = 4.25KK459 pKa = 11.39EE460 pKa = 4.61LDD462 pKa = 3.34QYY464 pKa = 11.17MATTKK469 pKa = 10.12TEE471 pKa = 3.65HH472 pKa = 6.1EE473 pKa = 3.96MDD475 pKa = 3.8YY476 pKa = 11.27LLKK479 pKa = 10.9NN480 pKa = 3.63
MM1 pKa = 7.21MSSTLSKK8 pKa = 10.35IHH10 pKa = 5.38VTNSTGLSLNEE21 pKa = 3.83RR22 pKa = 11.84FTAISTQPKK31 pKa = 8.77VVTNIGATRR40 pKa = 11.84RR41 pKa = 11.84SRR43 pKa = 11.84SRR45 pKa = 11.84SRR47 pKa = 11.84SVQRR51 pKa = 11.84QVPVAAAAEE60 pKa = 4.73RR61 pKa = 11.84ITSRR65 pKa = 11.84ANQRR69 pKa = 11.84LLAQLQRR76 pKa = 11.84KK77 pKa = 8.82HH78 pKa = 6.1KK79 pKa = 9.9VQAALKK85 pKa = 9.19LKK87 pKa = 9.02RR88 pKa = 11.84RR89 pKa = 11.84SMRR92 pKa = 11.84TVGGRR97 pKa = 11.84SNGSGGANGRR107 pKa = 11.84IRR109 pKa = 11.84RR110 pKa = 11.84GTVKK114 pKa = 10.58AFRR117 pKa = 11.84VGANGKK123 pKa = 8.74PIRR126 pKa = 11.84TNSLTRR132 pKa = 11.84IATMNADD139 pKa = 4.4RR140 pKa = 11.84EE141 pKa = 4.48ITNNRR146 pKa = 11.84SRR148 pKa = 11.84PLRR151 pKa = 11.84RR152 pKa = 11.84SNSSNNLAGRR162 pKa = 11.84LGPRR166 pKa = 11.84RR167 pKa = 11.84PTVGGAAQRR176 pKa = 11.84VARR179 pKa = 11.84RR180 pKa = 11.84NIDD183 pKa = 3.38RR184 pKa = 11.84QRR186 pKa = 11.84GRR188 pKa = 11.84NNVGIRR194 pKa = 11.84GRR196 pKa = 11.84STGINNRR203 pKa = 11.84GRR205 pKa = 11.84SRR207 pKa = 11.84SRR209 pKa = 11.84SRR211 pKa = 11.84TRR213 pKa = 11.84TLSDD217 pKa = 2.75NGRR220 pKa = 11.84NRR222 pKa = 11.84SRR224 pKa = 11.84TRR226 pKa = 11.84NNTDD230 pKa = 2.65NQLNNRR236 pKa = 11.84SRR238 pKa = 11.84SRR240 pKa = 11.84NRR242 pKa = 11.84NSANIQTNNNNNNVRR257 pKa = 11.84RR258 pKa = 11.84GRR260 pKa = 11.84SRR262 pKa = 11.84SRR264 pKa = 11.84SRR266 pKa = 11.84GRR268 pKa = 11.84AAVGSVNKK276 pKa = 10.54SNLPIKK282 pKa = 10.41ARR284 pKa = 11.84LGVRR288 pKa = 11.84PGVTANRR295 pKa = 11.84GTNNNRR301 pKa = 11.84RR302 pKa = 11.84GRR304 pKa = 11.84NASLTRR310 pKa = 11.84WGVQTGRR317 pKa = 11.84VQKK320 pKa = 10.39RR321 pKa = 11.84RR322 pKa = 11.84NSTNVNAAGNDD333 pKa = 3.42NNGRR337 pKa = 11.84GRR339 pKa = 11.84MRR341 pKa = 11.84SRR343 pKa = 11.84SAVRR347 pKa = 11.84NNSNNGNVRR356 pKa = 11.84SRR358 pKa = 11.84SRR360 pKa = 11.84SRR362 pKa = 11.84TRR364 pKa = 11.84KK365 pKa = 9.23GQVGQQRR372 pKa = 11.84QQQGSGQVRR381 pKa = 11.84QQRR384 pKa = 11.84NNRR387 pKa = 11.84APRR390 pKa = 11.84GRR392 pKa = 11.84QQQRR396 pKa = 11.84RR397 pKa = 11.84GNNTGNGQRR406 pKa = 11.84GTNNNGKK413 pKa = 9.07AQTQQRR419 pKa = 11.84RR420 pKa = 11.84GRR422 pKa = 11.84SRR424 pKa = 11.84SRR426 pKa = 11.84TGRR429 pKa = 11.84GANSGGNSRR438 pKa = 11.84NQGNKK443 pKa = 9.04SAAATKK449 pKa = 9.88PAVNKK454 pKa = 10.21DD455 pKa = 3.63ALDD458 pKa = 4.25KK459 pKa = 11.39EE460 pKa = 4.61LDD462 pKa = 3.34QYY464 pKa = 11.17MATTKK469 pKa = 10.12TEE471 pKa = 3.65HH472 pKa = 6.1EE473 pKa = 3.96MDD475 pKa = 3.8YY476 pKa = 11.27LLKK479 pKa = 10.9NN480 pKa = 3.63
Molecular weight: 53.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
7420241 |
30 |
19217 |
517.0 |
58.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.249 ± 0.024 | 1.914 ± 0.028 |
5.18 ± 0.021 | 6.492 ± 0.04 |
3.761 ± 0.02 | 5.069 ± 0.026 |
2.604 ± 0.013 | 5.815 ± 0.021 |
6.594 ± 0.032 | 8.867 ± 0.033 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.3 ± 0.012 | 6.209 ± 0.028 |
4.87 ± 0.028 | 4.941 ± 0.03 |
4.607 ± 0.019 | 8.344 ± 0.035 |
6.51 ± 0.03 | 5.545 ± 0.022 |
0.909 ± 0.008 | 3.212 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
