
Paraphaeosphaeria sporulosa
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; dothideomyceta; Dothideomycetes; Pleosporomycetidae; Pleosporales; Massarineae; Didymosphaeriaceae; Paraphaeosphaeria
Average proteome isoelectric point is 6.71
Get precalculated fractions of proteins
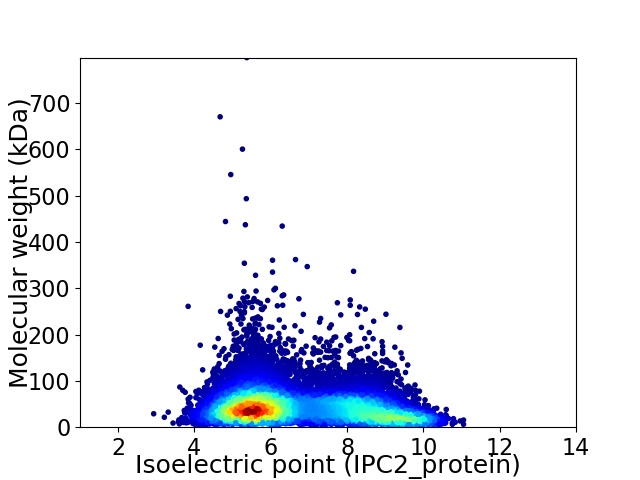
Virtual 2D-PAGE plot for 14732 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
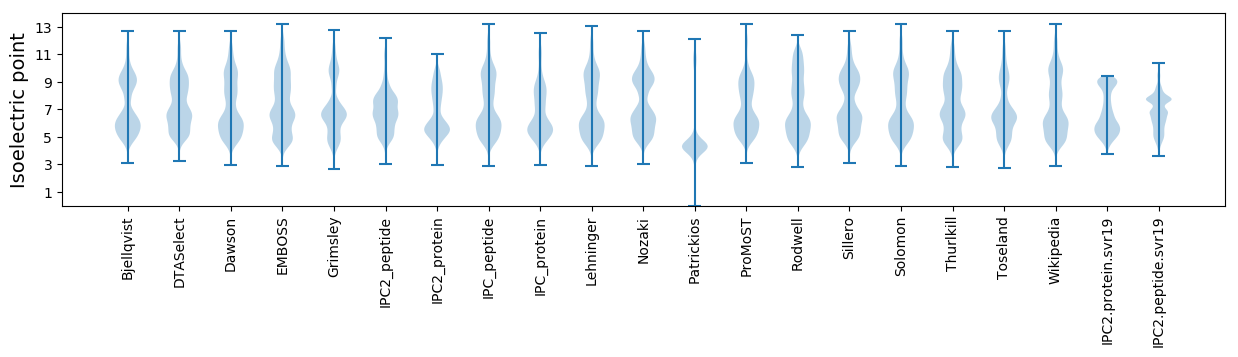
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A177CH65|A0A177CH65_9PLEO Alkaline phosphatase OS=Paraphaeosphaeria sporulosa OX=1460663 GN=CC84DRAFT_1090178 PE=3 SV=1
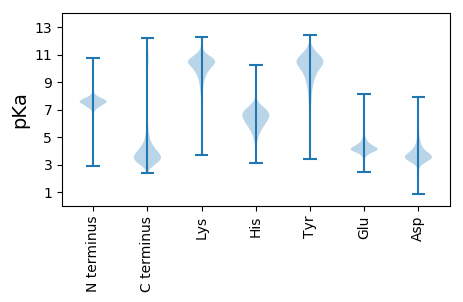
MM1 pKa = 7.79AYY3 pKa = 7.9ITLFSSLLPATLFFSPAQAADD24 pKa = 3.75CNLAQSRR31 pKa = 11.84RR32 pKa = 11.84SGDD35 pKa = 3.32PSGAAIASDD44 pKa = 5.36LRR46 pKa = 11.84GQDD49 pKa = 3.15NALLQSVCNGGFPPGSDD66 pKa = 5.17IISTWNTGSQIYY78 pKa = 10.4NVTRR82 pKa = 11.84FDD84 pKa = 3.55SSQPLQYY91 pKa = 10.96CFDD94 pKa = 4.04AFDD97 pKa = 5.11NIITQCIEE105 pKa = 3.54NDD107 pKa = 3.86NYY109 pKa = 10.78WGGDD113 pKa = 2.92WKK115 pKa = 10.99LGNEE119 pKa = 4.14IYY121 pKa = 10.5AIYY124 pKa = 10.52DD125 pKa = 3.75STWPEE130 pKa = 3.73HH131 pKa = 5.45VLEE134 pKa = 5.52AEE136 pKa = 4.42LEE138 pKa = 4.41SPPSSSAQDD147 pKa = 3.37VSPSSTPDD155 pKa = 3.07PPARR159 pKa = 11.84ATVVTTTIDD168 pKa = 3.5GFPITQTFVPTTIADD183 pKa = 4.1ASPTTSSTTVDD194 pKa = 3.54GVVVPLIIGAGGVAWIPFVPVGGAPPAVITPPADD228 pKa = 3.71LPDD231 pKa = 4.29GGNGDD236 pKa = 4.23GDD238 pKa = 4.85DD239 pKa = 4.43NSDD242 pKa = 3.84SQSSAPEE249 pKa = 4.16STSSSSSSSSSRR261 pKa = 11.84ALRR264 pKa = 11.84APDD267 pKa = 3.9DD268 pKa = 4.13VFPADD273 pKa = 3.88AGTATFDD280 pKa = 3.45SGAAAQIVFFGAAAPTPTPSSSSEE304 pKa = 3.59ASAEE308 pKa = 3.92PTVDD312 pKa = 3.52SNAEE316 pKa = 3.96AKK318 pKa = 10.07KK319 pKa = 10.44LCHH322 pKa = 5.98EE323 pKa = 4.52RR324 pKa = 11.84CGSKK328 pKa = 10.38PGLTTSDD335 pKa = 4.01GQWCLANCGCFLGAAGCC352 pKa = 4.03
MM1 pKa = 7.79AYY3 pKa = 7.9ITLFSSLLPATLFFSPAQAADD24 pKa = 3.75CNLAQSRR31 pKa = 11.84RR32 pKa = 11.84SGDD35 pKa = 3.32PSGAAIASDD44 pKa = 5.36LRR46 pKa = 11.84GQDD49 pKa = 3.15NALLQSVCNGGFPPGSDD66 pKa = 5.17IISTWNTGSQIYY78 pKa = 10.4NVTRR82 pKa = 11.84FDD84 pKa = 3.55SSQPLQYY91 pKa = 10.96CFDD94 pKa = 4.04AFDD97 pKa = 5.11NIITQCIEE105 pKa = 3.54NDD107 pKa = 3.86NYY109 pKa = 10.78WGGDD113 pKa = 2.92WKK115 pKa = 10.99LGNEE119 pKa = 4.14IYY121 pKa = 10.5AIYY124 pKa = 10.52DD125 pKa = 3.75STWPEE130 pKa = 3.73HH131 pKa = 5.45VLEE134 pKa = 5.52AEE136 pKa = 4.42LEE138 pKa = 4.41SPPSSSAQDD147 pKa = 3.37VSPSSTPDD155 pKa = 3.07PPARR159 pKa = 11.84ATVVTTTIDD168 pKa = 3.5GFPITQTFVPTTIADD183 pKa = 4.1ASPTTSSTTVDD194 pKa = 3.54GVVVPLIIGAGGVAWIPFVPVGGAPPAVITPPADD228 pKa = 3.71LPDD231 pKa = 4.29GGNGDD236 pKa = 4.23GDD238 pKa = 4.85DD239 pKa = 4.43NSDD242 pKa = 3.84SQSSAPEE249 pKa = 4.16STSSSSSSSSSRR261 pKa = 11.84ALRR264 pKa = 11.84APDD267 pKa = 3.9DD268 pKa = 4.13VFPADD273 pKa = 3.88AGTATFDD280 pKa = 3.45SGAAAQIVFFGAAAPTPTPSSSSEE304 pKa = 3.59ASAEE308 pKa = 3.92PTVDD312 pKa = 3.52SNAEE316 pKa = 3.96AKK318 pKa = 10.07KK319 pKa = 10.44LCHH322 pKa = 5.98EE323 pKa = 4.52RR324 pKa = 11.84CGSKK328 pKa = 10.38PGLTTSDD335 pKa = 4.01GQWCLANCGCFLGAAGCC352 pKa = 4.03
Molecular weight: 36.11 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A177C192|A0A177C192_9PLEO Uncharacterized protein OS=Paraphaeosphaeria sporulosa OX=1460663 GN=CC84DRAFT_1263047 PE=4 SV=1
MM1 pKa = 7.74PPMPKK6 pKa = 9.43RR7 pKa = 11.84RR8 pKa = 11.84GHH10 pKa = 5.51SFPRR14 pKa = 11.84LKK16 pKa = 10.56SKK18 pKa = 10.09PRR20 pKa = 11.84RR21 pKa = 11.84PSLPPSPPRR30 pKa = 11.84RR31 pKa = 11.84TKK33 pKa = 10.9LSTPSIRR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84SSWTASWLRR51 pKa = 11.84SSKK54 pKa = 9.64SWMPRR59 pKa = 11.84RR60 pKa = 11.84PATAKK65 pKa = 10.38LSKK68 pKa = 10.16SYY70 pKa = 9.22QARR73 pKa = 11.84TRR75 pKa = 11.84STPSCRR81 pKa = 11.84KK82 pKa = 7.66ITLLVVSKK90 pKa = 11.38AMVTRR95 pKa = 11.84PAFRR99 pKa = 11.84SWKK102 pKa = 9.35PQLQALNRR110 pKa = 11.84PRR112 pKa = 11.84VMFPAKK118 pKa = 10.02SQHH121 pKa = 6.13CKK123 pKa = 10.3SLSLRR128 pKa = 11.84RR129 pKa = 11.84RR130 pKa = 11.84RR131 pKa = 11.84LL132 pKa = 3.15
MM1 pKa = 7.74PPMPKK6 pKa = 9.43RR7 pKa = 11.84RR8 pKa = 11.84GHH10 pKa = 5.51SFPRR14 pKa = 11.84LKK16 pKa = 10.56SKK18 pKa = 10.09PRR20 pKa = 11.84RR21 pKa = 11.84PSLPPSPPRR30 pKa = 11.84RR31 pKa = 11.84TKK33 pKa = 10.9LSTPSIRR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84SSWTASWLRR51 pKa = 11.84SSKK54 pKa = 9.64SWMPRR59 pKa = 11.84RR60 pKa = 11.84PATAKK65 pKa = 10.38LSKK68 pKa = 10.16SYY70 pKa = 9.22QARR73 pKa = 11.84TRR75 pKa = 11.84STPSCRR81 pKa = 11.84KK82 pKa = 7.66ITLLVVSKK90 pKa = 11.38AMVTRR95 pKa = 11.84PAFRR99 pKa = 11.84SWKK102 pKa = 9.35PQLQALNRR110 pKa = 11.84PRR112 pKa = 11.84VMFPAKK118 pKa = 10.02SQHH121 pKa = 6.13CKK123 pKa = 10.3SLSLRR128 pKa = 11.84RR129 pKa = 11.84RR130 pKa = 11.84RR131 pKa = 11.84LL132 pKa = 3.15
Molecular weight: 15.41 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6378640 |
49 |
7206 |
433.0 |
47.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.051 ± 0.016 | 1.316 ± 0.009 |
5.531 ± 0.015 | 5.984 ± 0.019 |
3.746 ± 0.013 | 6.976 ± 0.021 |
2.473 ± 0.009 | 4.775 ± 0.013 |
4.883 ± 0.018 | 8.747 ± 0.021 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.166 ± 0.008 | 3.623 ± 0.011 |
6.108 ± 0.024 | 3.93 ± 0.015 |
6.11 ± 0.019 | 7.948 ± 0.022 |
6.054 ± 0.013 | 6.181 ± 0.016 |
1.567 ± 0.008 | 2.831 ± 0.01 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
