
Methanomassiliicoccus intestinalis (strain Issoire-Mx1)
Taxonomy: cellular organisms; Archaea; Candidatus Thermoplasmatota; Thermoplasmata; Methanomassiliicoccales; Methanomassiliicoccaceae; Methanomassiliicoccus; Candidatus Methanomassiliicoccus intestinalis
Average proteome isoelectric point is 5.78
Get precalculated fractions of proteins
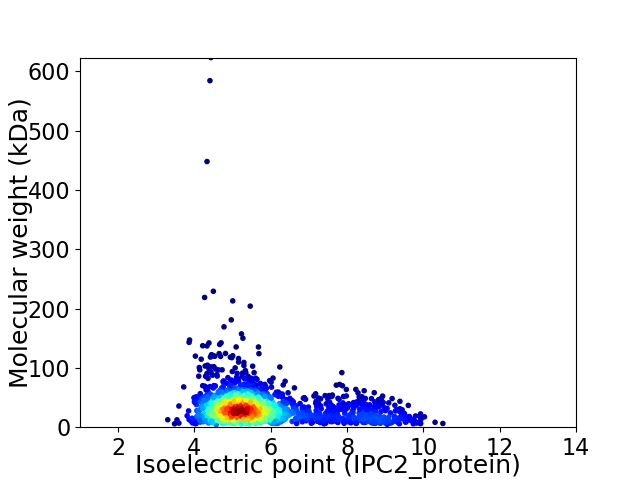
Virtual 2D-PAGE plot for 1826 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R9T7M1|R9T7M1_METII Translation initiation factor 5A OS=Methanomassiliicoccus intestinalis (strain Issoire-Mx1) OX=1295009 GN=eif5a PE=3 SV=1
MM1 pKa = 7.57NDD3 pKa = 2.86KK4 pKa = 10.76SKK6 pKa = 11.29NKK8 pKa = 10.07SIIMALAVFIAIAAFASPLLMSDD31 pKa = 5.05DD32 pKa = 4.37NGDD35 pKa = 3.68SNTVVLSDD43 pKa = 3.74EE44 pKa = 4.44APDD47 pKa = 3.82YY48 pKa = 11.5VSVSSQDD55 pKa = 4.63DD56 pKa = 3.27IDD58 pKa = 3.95NAAEE62 pKa = 4.01NSKK65 pKa = 10.79LLLNDD70 pKa = 4.44DD71 pKa = 4.08FTGSLTIPSGTSLYY85 pKa = 10.99GIDD88 pKa = 3.88IPKK91 pKa = 10.6DD92 pKa = 3.3LTDD95 pKa = 4.86LVGAPDD101 pKa = 3.83GAKK104 pKa = 9.9TITGDD109 pKa = 3.36VTLEE113 pKa = 3.98AGATLNFIKK122 pKa = 10.63INGKK126 pKa = 9.45VSICDD131 pKa = 3.41NNVTVNDD138 pKa = 3.7STATEE143 pKa = 3.75IRR145 pKa = 11.84YY146 pKa = 7.84CTINSEE152 pKa = 4.01TAIAISYY159 pKa = 10.88LDD161 pKa = 3.56IQAGDD166 pKa = 3.31ISIVSNEE173 pKa = 3.82INNSNTTNAILSFEE187 pKa = 4.29GARR190 pKa = 11.84DD191 pKa = 3.7GKK193 pKa = 10.69RR194 pKa = 11.84GATASEE200 pKa = 4.95SIFSLSISNNKK211 pKa = 9.63LGADD215 pKa = 3.54EE216 pKa = 5.1GRR218 pKa = 11.84AFSGNDD224 pKa = 3.22KK225 pKa = 11.02VGLSYY230 pKa = 11.22AGGSQISISNNIIADD245 pKa = 3.79APSWGASIMLRR256 pKa = 11.84SLNEE260 pKa = 3.4NSTFDD265 pKa = 3.45VSKK268 pKa = 11.53NNIANGDD275 pKa = 3.75KK276 pKa = 11.01PEE278 pKa = 4.16VGLSGGNAFDD288 pKa = 4.43GDD290 pKa = 4.29ANSFLDD296 pKa = 5.36GFTTDD301 pKa = 3.81AKK303 pKa = 11.14SLIIPIGSIAVLSEE317 pKa = 3.63GSEE320 pKa = 4.33FEE322 pKa = 4.19FEE324 pKa = 4.46RR325 pKa = 11.84ITVNGTITYY334 pKa = 9.76DD335 pKa = 3.21GSSISNVDD343 pKa = 3.73GISAIKK349 pKa = 10.35AGSLIINGEE358 pKa = 4.18LTPIGADD365 pKa = 3.36TPIVVSGDD373 pKa = 2.88ITLEE377 pKa = 3.99GSLMGKK383 pKa = 10.05LIILPEE389 pKa = 4.58EE390 pKa = 3.93IDD392 pKa = 3.27EE393 pKa = 4.92DD394 pKa = 4.77GNVLVKK400 pKa = 11.09SNIKK404 pKa = 9.97LRR406 pKa = 11.84NFEE409 pKa = 4.29TTDD412 pKa = 3.44GAVIEE417 pKa = 4.52YY418 pKa = 10.24KK419 pKa = 10.17LTVDD423 pKa = 4.92DD424 pKa = 4.98EE425 pKa = 5.28GSVSDD430 pKa = 4.56IGTISNVSEE439 pKa = 4.01IKK441 pKa = 10.01TNEE444 pKa = 3.75AGEE447 pKa = 4.36DD448 pKa = 3.67VVISNLYY455 pKa = 8.73EE456 pKa = 4.08DD457 pKa = 4.09RR458 pKa = 11.84GVIIDD463 pKa = 3.66VHH465 pKa = 7.79DD466 pKa = 3.42IEE468 pKa = 5.21YY469 pKa = 10.8NSEE472 pKa = 4.01YY473 pKa = 10.33QDD475 pKa = 3.65APVDD479 pKa = 3.79VTQFKK484 pKa = 10.26AANNIKK490 pKa = 9.43VLKK493 pKa = 9.86EE494 pKa = 3.36ITYY497 pKa = 10.22TNVCGAKK504 pKa = 9.9FVYY507 pKa = 10.2NVEE510 pKa = 5.04DD511 pKa = 4.16YY512 pKa = 9.19FTDD515 pKa = 4.41SILDD519 pKa = 3.59VNGISAKK526 pKa = 10.52VEE528 pKa = 4.53TIDD531 pKa = 4.11KK532 pKa = 10.3TSAFLNLKK540 pKa = 7.54VTNAGKK546 pKa = 10.79YY547 pKa = 7.79EE548 pKa = 3.85LTYY551 pKa = 10.37IVSTNAISDD560 pKa = 3.68DD561 pKa = 3.52GKK563 pKa = 10.81NIVDD567 pKa = 3.59SDD569 pKa = 4.0FKK571 pKa = 11.79VEE573 pKa = 4.05FTVSQKK579 pKa = 10.22VIKK582 pKa = 10.07YY583 pKa = 9.2ALPLVVKK590 pKa = 10.38IFDD593 pKa = 3.58GTTDD597 pKa = 3.03VFGNIVCLDD606 pKa = 3.41IFDD609 pKa = 4.32EE610 pKa = 4.95DD611 pKa = 3.77EE612 pKa = 4.31VSFLLEE618 pKa = 4.31FEE620 pKa = 4.53SADD623 pKa = 3.57AGIHH627 pKa = 5.35KK628 pKa = 10.58LIFKK632 pKa = 10.69GLDD635 pKa = 3.18GEE637 pKa = 4.78DD638 pKa = 3.13AGNYY642 pKa = 9.11ILADD646 pKa = 3.48YY647 pKa = 10.75YY648 pKa = 11.1FITNYY653 pKa = 8.75THH655 pKa = 7.34DD656 pKa = 3.91CCEE659 pKa = 3.85NSEE662 pKa = 4.16SCEE665 pKa = 4.19CGDD668 pKa = 5.44LPFSMDD674 pKa = 4.52DD675 pKa = 3.75CTCEE679 pKa = 5.32NDD681 pKa = 3.1FCMCKK686 pKa = 10.45FIFYY690 pKa = 10.98GLILPKK696 pKa = 10.19TITSDD701 pKa = 3.28MITISNVVPSFDD713 pKa = 4.71HH714 pKa = 7.0PDD716 pKa = 3.19KK717 pKa = 11.01VYY719 pKa = 8.69PTITVEE725 pKa = 4.33LLDD728 pKa = 4.07GTTVEE733 pKa = 5.24LDD735 pKa = 3.23TSKK738 pKa = 10.58MNEE741 pKa = 3.93RR742 pKa = 11.84DD743 pKa = 3.11YY744 pKa = 11.11DD745 pKa = 3.6IPFDD749 pKa = 3.78GFFVNKK755 pKa = 9.53NDD757 pKa = 3.47CEE759 pKa = 4.35QYY761 pKa = 11.07FEE763 pKa = 4.66FEE765 pKa = 4.79DD766 pKa = 5.09GISINDD772 pKa = 3.48AGDD775 pKa = 3.69YY776 pKa = 10.34YY777 pKa = 11.61AVICNHH783 pKa = 6.83PLAPSYY789 pKa = 11.28NMNYY793 pKa = 9.85SSSFSGSNYY802 pKa = 8.59IAGPILEE809 pKa = 4.74NIFYY813 pKa = 10.3EE814 pKa = 4.37YY815 pKa = 10.11IEE817 pKa = 4.01NVIYY821 pKa = 9.8EE822 pKa = 4.69LPDD825 pKa = 3.48ATDD828 pKa = 3.11NGMDD832 pKa = 3.73NSIPMYY838 pKa = 9.96LTDD841 pKa = 5.99LLTQFISHH849 pKa = 6.5FTNNGEE855 pKa = 4.31SVNIQDD861 pKa = 5.37IINLLFTLPINGEE874 pKa = 3.98DD875 pKa = 3.73SGNKK879 pKa = 9.99AIVDD883 pKa = 3.22IDD885 pKa = 3.79DD886 pKa = 3.54RR887 pKa = 11.84TLFMMYY893 pKa = 9.99YY894 pKa = 10.01LASAEE899 pKa = 4.12EE900 pKa = 3.97NANLKK905 pKa = 10.94GEE907 pKa = 4.04LAYY910 pKa = 10.68DD911 pKa = 3.98GGNNIIRR918 pKa = 11.84FSEE921 pKa = 4.09DD922 pKa = 2.97LKK924 pKa = 11.58NEE926 pKa = 4.01AGLRR930 pKa = 11.84AWYY933 pKa = 10.33LSFNNQLSEE942 pKa = 4.23FGEE945 pKa = 4.4GDD947 pKa = 3.26YY948 pKa = 10.98HH949 pKa = 8.48LRR951 pKa = 11.84ISADD955 pKa = 3.19DD956 pKa = 3.7VTIAEE961 pKa = 4.36YY962 pKa = 10.19TFTFSSDD969 pKa = 3.71GIDD972 pKa = 4.45DD973 pKa = 4.2IVNVIYY979 pKa = 10.62DD980 pKa = 3.56EE981 pKa = 5.79GYY983 pKa = 11.36ALDD986 pKa = 5.26FANAWASKK994 pKa = 10.41KK995 pKa = 10.56GFEE998 pKa = 4.35SKK1000 pKa = 10.58SGIVINYY1007 pKa = 9.17HH1008 pKa = 6.23YY1009 pKa = 10.45IYY1011 pKa = 9.98DD1012 pKa = 4.2DD1013 pKa = 3.61KK1014 pKa = 11.41GSEE1017 pKa = 3.61IRR1019 pKa = 11.84EE1020 pKa = 3.82EE1021 pKa = 4.33HH1022 pKa = 6.42IEE1024 pKa = 3.99QIINIAMLKK1033 pKa = 10.52NGLQYY1038 pKa = 10.57TYY1040 pKa = 11.2SNVGAQLQQYY1050 pKa = 9.17EE1051 pKa = 4.42FLRR1054 pKa = 11.84WSTVGSPSLYY1064 pKa = 10.8DD1065 pKa = 3.69DD1066 pKa = 4.07ACLVDD1071 pKa = 4.73NEE1073 pKa = 4.73LFHH1076 pKa = 7.27FNALEE1081 pKa = 4.37VIDD1084 pKa = 4.1RR1085 pKa = 11.84TDD1087 pKa = 3.46LEE1089 pKa = 4.68SYY1091 pKa = 10.91AVDD1094 pKa = 3.6GVLNLYY1100 pKa = 10.12AVYY1103 pKa = 10.92DD1104 pKa = 3.97MKK1106 pKa = 10.95TPKK1109 pKa = 9.7PDD1111 pKa = 3.17NKK1113 pKa = 10.95SIDD1116 pKa = 3.79NVAAFYY1122 pKa = 10.78DD1123 pKa = 3.55IDD1125 pKa = 3.82AKK1127 pKa = 10.69KK1128 pKa = 10.7VQDD1131 pKa = 3.75LMSLYY1136 pKa = 10.29GINVDD1141 pKa = 4.18DD1142 pKa = 4.74VISRR1146 pKa = 11.84TMAFIYY1152 pKa = 8.99EE1153 pKa = 3.88QSGYY1157 pKa = 11.43SNTTMKK1163 pKa = 10.95GILYY1167 pKa = 9.99KK1168 pKa = 10.69LVNGEE1173 pKa = 4.04LLEE1176 pKa = 4.65VYY1178 pKa = 10.53HH1179 pKa = 7.02EE1180 pKa = 5.39DD1181 pKa = 4.7NLNSDD1186 pKa = 3.81DD1187 pKa = 5.94GLRR1190 pKa = 11.84AWYY1193 pKa = 9.43FSFDD1197 pKa = 3.74DD1198 pKa = 3.96QVNSAVDD1205 pKa = 3.57TYY1207 pKa = 11.71GLAGYY1212 pKa = 10.18YY1213 pKa = 9.58DD1214 pKa = 3.88ARR1216 pKa = 11.84IVAVDD1221 pKa = 3.36PEE1223 pKa = 4.52GNEE1226 pKa = 4.19TEE1228 pKa = 4.07LLSAKK1233 pKa = 9.73FHH1235 pKa = 6.41IAEE1238 pKa = 4.29EE1239 pKa = 4.6LYY1241 pKa = 10.71GANITVDD1248 pKa = 3.79YY1249 pKa = 11.43VNVDD1253 pKa = 4.44DD1254 pKa = 5.86INDD1257 pKa = 3.57AVKK1260 pKa = 10.81VYY1262 pKa = 10.4IEE1264 pKa = 4.51PKK1266 pKa = 10.2NGCLVPAGDD1275 pKa = 5.34LIVAYY1280 pKa = 10.47SFMSSGPFGDD1290 pKa = 4.42EE1291 pKa = 3.87PASALADD1298 pKa = 3.41AVPIPVSSSTQIIDD1312 pKa = 3.35VDD1314 pKa = 3.46ISGMDD1319 pKa = 3.39SANAIFKK1326 pKa = 9.85WIDD1329 pKa = 3.38SSGHH1333 pKa = 4.33TVYY1336 pKa = 11.03SLSNTVTSQQ1345 pKa = 2.73
MM1 pKa = 7.57NDD3 pKa = 2.86KK4 pKa = 10.76SKK6 pKa = 11.29NKK8 pKa = 10.07SIIMALAVFIAIAAFASPLLMSDD31 pKa = 5.05DD32 pKa = 4.37NGDD35 pKa = 3.68SNTVVLSDD43 pKa = 3.74EE44 pKa = 4.44APDD47 pKa = 3.82YY48 pKa = 11.5VSVSSQDD55 pKa = 4.63DD56 pKa = 3.27IDD58 pKa = 3.95NAAEE62 pKa = 4.01NSKK65 pKa = 10.79LLLNDD70 pKa = 4.44DD71 pKa = 4.08FTGSLTIPSGTSLYY85 pKa = 10.99GIDD88 pKa = 3.88IPKK91 pKa = 10.6DD92 pKa = 3.3LTDD95 pKa = 4.86LVGAPDD101 pKa = 3.83GAKK104 pKa = 9.9TITGDD109 pKa = 3.36VTLEE113 pKa = 3.98AGATLNFIKK122 pKa = 10.63INGKK126 pKa = 9.45VSICDD131 pKa = 3.41NNVTVNDD138 pKa = 3.7STATEE143 pKa = 3.75IRR145 pKa = 11.84YY146 pKa = 7.84CTINSEE152 pKa = 4.01TAIAISYY159 pKa = 10.88LDD161 pKa = 3.56IQAGDD166 pKa = 3.31ISIVSNEE173 pKa = 3.82INNSNTTNAILSFEE187 pKa = 4.29GARR190 pKa = 11.84DD191 pKa = 3.7GKK193 pKa = 10.69RR194 pKa = 11.84GATASEE200 pKa = 4.95SIFSLSISNNKK211 pKa = 9.63LGADD215 pKa = 3.54EE216 pKa = 5.1GRR218 pKa = 11.84AFSGNDD224 pKa = 3.22KK225 pKa = 11.02VGLSYY230 pKa = 11.22AGGSQISISNNIIADD245 pKa = 3.79APSWGASIMLRR256 pKa = 11.84SLNEE260 pKa = 3.4NSTFDD265 pKa = 3.45VSKK268 pKa = 11.53NNIANGDD275 pKa = 3.75KK276 pKa = 11.01PEE278 pKa = 4.16VGLSGGNAFDD288 pKa = 4.43GDD290 pKa = 4.29ANSFLDD296 pKa = 5.36GFTTDD301 pKa = 3.81AKK303 pKa = 11.14SLIIPIGSIAVLSEE317 pKa = 3.63GSEE320 pKa = 4.33FEE322 pKa = 4.19FEE324 pKa = 4.46RR325 pKa = 11.84ITVNGTITYY334 pKa = 9.76DD335 pKa = 3.21GSSISNVDD343 pKa = 3.73GISAIKK349 pKa = 10.35AGSLIINGEE358 pKa = 4.18LTPIGADD365 pKa = 3.36TPIVVSGDD373 pKa = 2.88ITLEE377 pKa = 3.99GSLMGKK383 pKa = 10.05LIILPEE389 pKa = 4.58EE390 pKa = 3.93IDD392 pKa = 3.27EE393 pKa = 4.92DD394 pKa = 4.77GNVLVKK400 pKa = 11.09SNIKK404 pKa = 9.97LRR406 pKa = 11.84NFEE409 pKa = 4.29TTDD412 pKa = 3.44GAVIEE417 pKa = 4.52YY418 pKa = 10.24KK419 pKa = 10.17LTVDD423 pKa = 4.92DD424 pKa = 4.98EE425 pKa = 5.28GSVSDD430 pKa = 4.56IGTISNVSEE439 pKa = 4.01IKK441 pKa = 10.01TNEE444 pKa = 3.75AGEE447 pKa = 4.36DD448 pKa = 3.67VVISNLYY455 pKa = 8.73EE456 pKa = 4.08DD457 pKa = 4.09RR458 pKa = 11.84GVIIDD463 pKa = 3.66VHH465 pKa = 7.79DD466 pKa = 3.42IEE468 pKa = 5.21YY469 pKa = 10.8NSEE472 pKa = 4.01YY473 pKa = 10.33QDD475 pKa = 3.65APVDD479 pKa = 3.79VTQFKK484 pKa = 10.26AANNIKK490 pKa = 9.43VLKK493 pKa = 9.86EE494 pKa = 3.36ITYY497 pKa = 10.22TNVCGAKK504 pKa = 9.9FVYY507 pKa = 10.2NVEE510 pKa = 5.04DD511 pKa = 4.16YY512 pKa = 9.19FTDD515 pKa = 4.41SILDD519 pKa = 3.59VNGISAKK526 pKa = 10.52VEE528 pKa = 4.53TIDD531 pKa = 4.11KK532 pKa = 10.3TSAFLNLKK540 pKa = 7.54VTNAGKK546 pKa = 10.79YY547 pKa = 7.79EE548 pKa = 3.85LTYY551 pKa = 10.37IVSTNAISDD560 pKa = 3.68DD561 pKa = 3.52GKK563 pKa = 10.81NIVDD567 pKa = 3.59SDD569 pKa = 4.0FKK571 pKa = 11.79VEE573 pKa = 4.05FTVSQKK579 pKa = 10.22VIKK582 pKa = 10.07YY583 pKa = 9.2ALPLVVKK590 pKa = 10.38IFDD593 pKa = 3.58GTTDD597 pKa = 3.03VFGNIVCLDD606 pKa = 3.41IFDD609 pKa = 4.32EE610 pKa = 4.95DD611 pKa = 3.77EE612 pKa = 4.31VSFLLEE618 pKa = 4.31FEE620 pKa = 4.53SADD623 pKa = 3.57AGIHH627 pKa = 5.35KK628 pKa = 10.58LIFKK632 pKa = 10.69GLDD635 pKa = 3.18GEE637 pKa = 4.78DD638 pKa = 3.13AGNYY642 pKa = 9.11ILADD646 pKa = 3.48YY647 pKa = 10.75YY648 pKa = 11.1FITNYY653 pKa = 8.75THH655 pKa = 7.34DD656 pKa = 3.91CCEE659 pKa = 3.85NSEE662 pKa = 4.16SCEE665 pKa = 4.19CGDD668 pKa = 5.44LPFSMDD674 pKa = 4.52DD675 pKa = 3.75CTCEE679 pKa = 5.32NDD681 pKa = 3.1FCMCKK686 pKa = 10.45FIFYY690 pKa = 10.98GLILPKK696 pKa = 10.19TITSDD701 pKa = 3.28MITISNVVPSFDD713 pKa = 4.71HH714 pKa = 7.0PDD716 pKa = 3.19KK717 pKa = 11.01VYY719 pKa = 8.69PTITVEE725 pKa = 4.33LLDD728 pKa = 4.07GTTVEE733 pKa = 5.24LDD735 pKa = 3.23TSKK738 pKa = 10.58MNEE741 pKa = 3.93RR742 pKa = 11.84DD743 pKa = 3.11YY744 pKa = 11.11DD745 pKa = 3.6IPFDD749 pKa = 3.78GFFVNKK755 pKa = 9.53NDD757 pKa = 3.47CEE759 pKa = 4.35QYY761 pKa = 11.07FEE763 pKa = 4.66FEE765 pKa = 4.79DD766 pKa = 5.09GISINDD772 pKa = 3.48AGDD775 pKa = 3.69YY776 pKa = 10.34YY777 pKa = 11.61AVICNHH783 pKa = 6.83PLAPSYY789 pKa = 11.28NMNYY793 pKa = 9.85SSSFSGSNYY802 pKa = 8.59IAGPILEE809 pKa = 4.74NIFYY813 pKa = 10.3EE814 pKa = 4.37YY815 pKa = 10.11IEE817 pKa = 4.01NVIYY821 pKa = 9.8EE822 pKa = 4.69LPDD825 pKa = 3.48ATDD828 pKa = 3.11NGMDD832 pKa = 3.73NSIPMYY838 pKa = 9.96LTDD841 pKa = 5.99LLTQFISHH849 pKa = 6.5FTNNGEE855 pKa = 4.31SVNIQDD861 pKa = 5.37IINLLFTLPINGEE874 pKa = 3.98DD875 pKa = 3.73SGNKK879 pKa = 9.99AIVDD883 pKa = 3.22IDD885 pKa = 3.79DD886 pKa = 3.54RR887 pKa = 11.84TLFMMYY893 pKa = 9.99YY894 pKa = 10.01LASAEE899 pKa = 4.12EE900 pKa = 3.97NANLKK905 pKa = 10.94GEE907 pKa = 4.04LAYY910 pKa = 10.68DD911 pKa = 3.98GGNNIIRR918 pKa = 11.84FSEE921 pKa = 4.09DD922 pKa = 2.97LKK924 pKa = 11.58NEE926 pKa = 4.01AGLRR930 pKa = 11.84AWYY933 pKa = 10.33LSFNNQLSEE942 pKa = 4.23FGEE945 pKa = 4.4GDD947 pKa = 3.26YY948 pKa = 10.98HH949 pKa = 8.48LRR951 pKa = 11.84ISADD955 pKa = 3.19DD956 pKa = 3.7VTIAEE961 pKa = 4.36YY962 pKa = 10.19TFTFSSDD969 pKa = 3.71GIDD972 pKa = 4.45DD973 pKa = 4.2IVNVIYY979 pKa = 10.62DD980 pKa = 3.56EE981 pKa = 5.79GYY983 pKa = 11.36ALDD986 pKa = 5.26FANAWASKK994 pKa = 10.41KK995 pKa = 10.56GFEE998 pKa = 4.35SKK1000 pKa = 10.58SGIVINYY1007 pKa = 9.17HH1008 pKa = 6.23YY1009 pKa = 10.45IYY1011 pKa = 9.98DD1012 pKa = 4.2DD1013 pKa = 3.61KK1014 pKa = 11.41GSEE1017 pKa = 3.61IRR1019 pKa = 11.84EE1020 pKa = 3.82EE1021 pKa = 4.33HH1022 pKa = 6.42IEE1024 pKa = 3.99QIINIAMLKK1033 pKa = 10.52NGLQYY1038 pKa = 10.57TYY1040 pKa = 11.2SNVGAQLQQYY1050 pKa = 9.17EE1051 pKa = 4.42FLRR1054 pKa = 11.84WSTVGSPSLYY1064 pKa = 10.8DD1065 pKa = 3.69DD1066 pKa = 4.07ACLVDD1071 pKa = 4.73NEE1073 pKa = 4.73LFHH1076 pKa = 7.27FNALEE1081 pKa = 4.37VIDD1084 pKa = 4.1RR1085 pKa = 11.84TDD1087 pKa = 3.46LEE1089 pKa = 4.68SYY1091 pKa = 10.91AVDD1094 pKa = 3.6GVLNLYY1100 pKa = 10.12AVYY1103 pKa = 10.92DD1104 pKa = 3.97MKK1106 pKa = 10.95TPKK1109 pKa = 9.7PDD1111 pKa = 3.17NKK1113 pKa = 10.95SIDD1116 pKa = 3.79NVAAFYY1122 pKa = 10.78DD1123 pKa = 3.55IDD1125 pKa = 3.82AKK1127 pKa = 10.69KK1128 pKa = 10.7VQDD1131 pKa = 3.75LMSLYY1136 pKa = 10.29GINVDD1141 pKa = 4.18DD1142 pKa = 4.74VISRR1146 pKa = 11.84TMAFIYY1152 pKa = 8.99EE1153 pKa = 3.88QSGYY1157 pKa = 11.43SNTTMKK1163 pKa = 10.95GILYY1167 pKa = 9.99KK1168 pKa = 10.69LVNGEE1173 pKa = 4.04LLEE1176 pKa = 4.65VYY1178 pKa = 10.53HH1179 pKa = 7.02EE1180 pKa = 5.39DD1181 pKa = 4.7NLNSDD1186 pKa = 3.81DD1187 pKa = 5.94GLRR1190 pKa = 11.84AWYY1193 pKa = 9.43FSFDD1197 pKa = 3.74DD1198 pKa = 3.96QVNSAVDD1205 pKa = 3.57TYY1207 pKa = 11.71GLAGYY1212 pKa = 10.18YY1213 pKa = 9.58DD1214 pKa = 3.88ARR1216 pKa = 11.84IVAVDD1221 pKa = 3.36PEE1223 pKa = 4.52GNEE1226 pKa = 4.19TEE1228 pKa = 4.07LLSAKK1233 pKa = 9.73FHH1235 pKa = 6.41IAEE1238 pKa = 4.29EE1239 pKa = 4.6LYY1241 pKa = 10.71GANITVDD1248 pKa = 3.79YY1249 pKa = 11.43VNVDD1253 pKa = 4.44DD1254 pKa = 5.86INDD1257 pKa = 3.57AVKK1260 pKa = 10.81VYY1262 pKa = 10.4IEE1264 pKa = 4.51PKK1266 pKa = 10.2NGCLVPAGDD1275 pKa = 5.34LIVAYY1280 pKa = 10.47SFMSSGPFGDD1290 pKa = 4.42EE1291 pKa = 3.87PASALADD1298 pKa = 3.41AVPIPVSSSTQIIDD1312 pKa = 3.35VDD1314 pKa = 3.46ISGMDD1319 pKa = 3.39SANAIFKK1326 pKa = 9.85WIDD1329 pKa = 3.38SSGHH1333 pKa = 4.33TVYY1336 pKa = 11.03SLSNTVTSQQ1345 pKa = 2.73
Molecular weight: 146.99 kDa
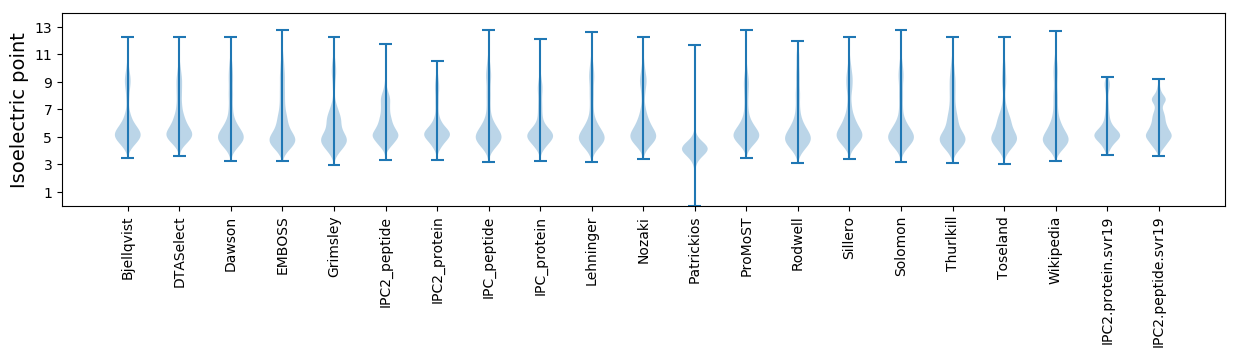
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R9TAI4|R9TAI4_METII 7-cyano-7-deazaguanine synthase OS=Methanomassiliicoccus intestinalis (strain Issoire-Mx1) OX=1295009 GN=queC PE=3 SV=1
MM1 pKa = 7.27ARR3 pKa = 11.84KK4 pKa = 8.58PNRR7 pKa = 11.84MYY9 pKa = 10.76RR10 pKa = 11.84QIRR13 pKa = 11.84GQAYY17 pKa = 6.57TRR19 pKa = 11.84RR20 pKa = 11.84EE21 pKa = 4.01YY22 pKa = 10.51MGGVPANRR30 pKa = 11.84ISTYY34 pKa = 11.06NLGNVTGDD42 pKa = 3.29FPIVMNLRR50 pKa = 11.84IKK52 pKa = 9.85EE53 pKa = 4.11PCQIRR58 pKa = 11.84HH59 pKa = 5.44TALEE63 pKa = 4.03AARR66 pKa = 11.84IAANRR71 pKa = 11.84ALSKK75 pKa = 10.01GTTSASYY82 pKa = 10.18HH83 pKa = 5.18LRR85 pKa = 11.84IRR87 pKa = 11.84LYY89 pKa = 7.95PHH91 pKa = 6.13IVLRR95 pKa = 11.84EE96 pKa = 3.92HH97 pKa = 7.26KK98 pKa = 9.02MATGAGADD106 pKa = 3.4RR107 pKa = 11.84VSGGMRR113 pKa = 11.84SAFGKK118 pKa = 10.64CVGTACRR125 pKa = 11.84VEE127 pKa = 4.35SGTAIITLRR136 pKa = 11.84VTPQTFQLGKK146 pKa = 9.24NALWKK151 pKa = 10.77ASMKK155 pKa = 10.57LPSPCQIEE163 pKa = 4.17IEE165 pKa = 4.21KK166 pKa = 10.68GAEE169 pKa = 3.78LVAA172 pKa = 5.36
MM1 pKa = 7.27ARR3 pKa = 11.84KK4 pKa = 8.58PNRR7 pKa = 11.84MYY9 pKa = 10.76RR10 pKa = 11.84QIRR13 pKa = 11.84GQAYY17 pKa = 6.57TRR19 pKa = 11.84RR20 pKa = 11.84EE21 pKa = 4.01YY22 pKa = 10.51MGGVPANRR30 pKa = 11.84ISTYY34 pKa = 11.06NLGNVTGDD42 pKa = 3.29FPIVMNLRR50 pKa = 11.84IKK52 pKa = 9.85EE53 pKa = 4.11PCQIRR58 pKa = 11.84HH59 pKa = 5.44TALEE63 pKa = 4.03AARR66 pKa = 11.84IAANRR71 pKa = 11.84ALSKK75 pKa = 10.01GTTSASYY82 pKa = 10.18HH83 pKa = 5.18LRR85 pKa = 11.84IRR87 pKa = 11.84LYY89 pKa = 7.95PHH91 pKa = 6.13IVLRR95 pKa = 11.84EE96 pKa = 3.92HH97 pKa = 7.26KK98 pKa = 9.02MATGAGADD106 pKa = 3.4RR107 pKa = 11.84VSGGMRR113 pKa = 11.84SAFGKK118 pKa = 10.64CVGTACRR125 pKa = 11.84VEE127 pKa = 4.35SGTAIITLRR136 pKa = 11.84VTPQTFQLGKK146 pKa = 9.24NALWKK151 pKa = 10.77ASMKK155 pKa = 10.57LPSPCQIEE163 pKa = 4.17IEE165 pKa = 4.21KK166 pKa = 10.68GAEE169 pKa = 3.78LVAA172 pKa = 5.36
Molecular weight: 18.96 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
562683 |
38 |
5834 |
308.2 |
34.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.396 ± 0.066 | 1.537 ± 0.041 |
6.217 ± 0.054 | 6.847 ± 0.076 |
3.727 ± 0.038 | 7.232 ± 0.091 |
1.626 ± 0.026 | 8.09 ± 0.054 |
6.455 ± 0.057 | 8.584 ± 0.078 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.912 ± 0.047 | 4.593 ± 0.083 |
3.721 ± 0.033 | 2.367 ± 0.031 |
4.01 ± 0.069 | 7.474 ± 0.068 |
5.5 ± 0.096 | 7.065 ± 0.047 |
0.94 ± 0.02 | 3.704 ± 0.054 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
