
Perca flavescens (American yellow perch) (Morone flavescens)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Actinopterygii; Actinopteri; Neopterygii; Teleostei; Osteoglossocephalai; Clupeocephala; Euteleosteomorpha; Neoteleostei; Eurypterygia; Ctenosquamata;
Average proteome isoelectric point is 6.51
Get precalculated fractions of proteins
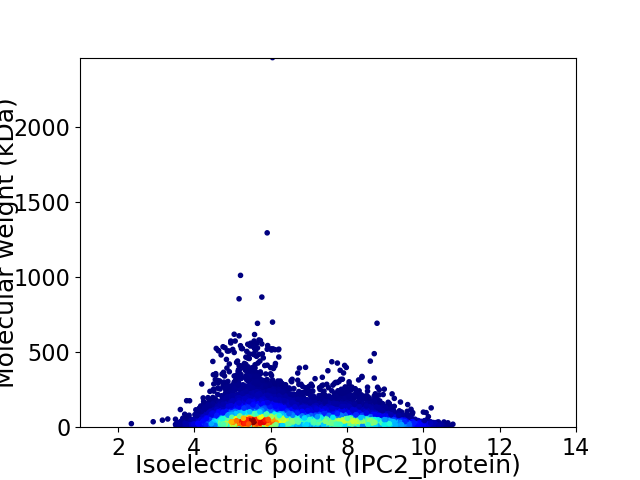
Virtual 2D-PAGE plot for 21644 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A484CST4|A0A484CST4_PERFV Uncharacterized protein OS=Perca flavescens OX=8167 GN=EPR50_G00139000 PE=4 SV=1
MM1 pKa = 7.49VDD3 pKa = 3.99FGEE6 pKa = 4.19ILTTIGDD13 pKa = 3.52FGFFQKK19 pKa = 10.94LILFGLTLPNVLLPVFFCSFLFIQSDD45 pKa = 4.09PEE47 pKa = 4.12RR48 pKa = 11.84HH49 pKa = 6.65CNTDD53 pKa = 3.05WILQADD59 pKa = 4.25SNLTTDD65 pKa = 3.81DD66 pKa = 4.02QLNLTLPRR74 pKa = 11.84EE75 pKa = 4.14EE76 pKa = 5.45DD77 pKa = 3.65GTFSRR82 pKa = 11.84CQMFVPVDD90 pKa = 3.33WDD92 pKa = 3.4VGDD95 pKa = 3.42IRR97 pKa = 11.84EE98 pKa = 4.18FGLNEE103 pKa = 3.75TTGCQNGWVYY113 pKa = 11.55YY114 pKa = 8.15NTLYY118 pKa = 10.7EE119 pKa = 4.13ATIVTDD125 pKa = 3.52VSEE128 pKa = 4.34DD129 pKa = 3.21AA130 pKa = 4.9
MM1 pKa = 7.49VDD3 pKa = 3.99FGEE6 pKa = 4.19ILTTIGDD13 pKa = 3.52FGFFQKK19 pKa = 10.94LILFGLTLPNVLLPVFFCSFLFIQSDD45 pKa = 4.09PEE47 pKa = 4.12RR48 pKa = 11.84HH49 pKa = 6.65CNTDD53 pKa = 3.05WILQADD59 pKa = 4.25SNLTTDD65 pKa = 3.81DD66 pKa = 4.02QLNLTLPRR74 pKa = 11.84EE75 pKa = 4.14EE76 pKa = 5.45DD77 pKa = 3.65GTFSRR82 pKa = 11.84CQMFVPVDD90 pKa = 3.33WDD92 pKa = 3.4VGDD95 pKa = 3.42IRR97 pKa = 11.84EE98 pKa = 4.18FGLNEE103 pKa = 3.75TTGCQNGWVYY113 pKa = 11.55YY114 pKa = 8.15NTLYY118 pKa = 10.7EE119 pKa = 4.13ATIVTDD125 pKa = 3.52VSEE128 pKa = 4.34DD129 pKa = 3.21AA130 pKa = 4.9
Molecular weight: 14.84 kDa
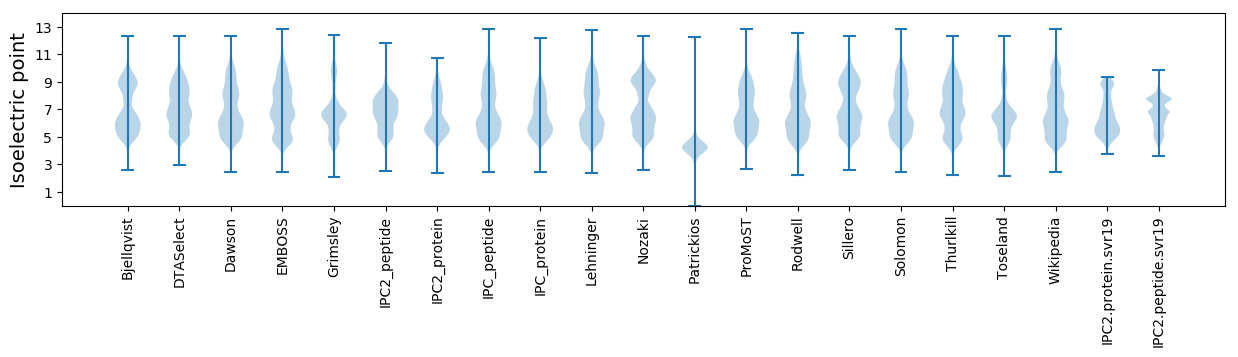
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A484DFF3|A0A484DFF3_PERFV Guanine nucleotide-binding protein-like 3 OS=Perca flavescens OX=8167 GN=EPR50_G00041740 PE=4 SV=1
MM1 pKa = 7.44GFLRR5 pKa = 11.84RR6 pKa = 11.84VAGVSLRR13 pKa = 11.84DD14 pKa = 3.38RR15 pKa = 11.84RR16 pKa = 11.84PQQRR20 pKa = 11.84RR21 pKa = 11.84KK22 pKa = 9.82QKK24 pKa = 10.56PNTPSTKK31 pKa = 10.11RR32 pKa = 11.84LTSQHH37 pKa = 5.78SAQGTSEE44 pKa = 4.44SKK46 pKa = 10.7AHH48 pKa = 6.44PLSPAEE54 pKa = 4.96DD55 pKa = 3.3KK56 pKa = 10.79MEE58 pKa = 4.86PKK60 pKa = 9.97VQPDD64 pKa = 3.82TQPCSHH70 pKa = 7.13RR71 pKa = 11.84EE72 pKa = 3.86HH73 pKa = 7.07KK74 pKa = 9.14PTGFRR79 pKa = 11.84VSRR82 pKa = 11.84HH83 pKa = 4.12TAAFSCHH90 pKa = 6.27RR91 pKa = 11.84LSDD94 pKa = 3.83SSPTLQEE101 pKa = 3.99SLSPKK106 pKa = 10.52AEE108 pKa = 3.94VGGLAGHH115 pKa = 6.82GEE117 pKa = 4.3GDD119 pKa = 3.45SDD121 pKa = 3.97TDD123 pKa = 3.6LSEE126 pKa = 4.44SEE128 pKa = 4.6RR129 pKa = 11.84LPVSPSGGVPPQLQLRR145 pKa = 11.84PEE147 pKa = 4.38VIEE150 pKa = 5.39AEE152 pKa = 4.31HH153 pKa = 6.55CPSRR157 pKa = 11.84SHH159 pKa = 7.25RR160 pKa = 11.84PRR162 pKa = 11.84GHH164 pKa = 5.57NHH166 pKa = 6.04GAFDD170 pKa = 4.99FPDD173 pKa = 4.44FLPPPFNSWSLSQLAVFYY191 pKa = 11.14NMEE194 pKa = 4.17GRR196 pKa = 11.84GGPRR200 pKa = 11.84PRR202 pKa = 11.84PMGPLEE208 pKa = 4.54RR209 pKa = 11.84YY210 pKa = 9.59LDD212 pKa = 3.76RR213 pKa = 11.84LLQLEE218 pKa = 4.24WHH220 pKa = 6.59QIQTVQGEE228 pKa = 4.66GRR230 pKa = 11.84KK231 pKa = 10.24SMGSDD236 pKa = 3.23VLSSCHH242 pKa = 6.44RR243 pKa = 11.84SHH245 pKa = 7.17AAASSRR251 pKa = 11.84LSSPKK256 pKa = 10.31CILQCQRR263 pKa = 11.84AFSLTFLSCLASHH276 pKa = 6.78SALLSGCACTLCRR289 pKa = 11.84IRR291 pKa = 11.84YY292 pKa = 5.9STCSTLCCRR301 pKa = 11.84SSHH304 pKa = 5.75HH305 pKa = 5.63THH307 pKa = 6.9HH308 pKa = 6.74SRR310 pKa = 11.84LSPMLEE316 pKa = 3.57RR317 pKa = 11.84RR318 pKa = 11.84GPTLLPKK325 pKa = 10.34RR326 pKa = 11.84SYY328 pKa = 11.11SEE330 pKa = 4.16NRR332 pKa = 11.84VKK334 pKa = 10.96SSDD337 pKa = 3.33RR338 pKa = 11.84SSAPRR343 pKa = 11.84SQVFSSPVRR352 pKa = 11.84TNSHH356 pKa = 6.07LRR358 pKa = 11.84RR359 pKa = 11.84MQASGNIRR367 pKa = 11.84NPVKK371 pKa = 10.49VANTKK376 pKa = 9.13PHH378 pKa = 4.9STARR382 pKa = 11.84GGSVGAGTDD391 pKa = 3.1RR392 pKa = 11.84VGALGDD398 pKa = 3.63VVDD401 pKa = 4.1YY402 pKa = 8.86RR403 pKa = 11.84TGGFRR408 pKa = 11.84RR409 pKa = 11.84RR410 pKa = 11.84SGSEE414 pKa = 3.34QRR416 pKa = 11.84RR417 pKa = 11.84GGVEE421 pKa = 3.85RR422 pKa = 11.84QQGASEE428 pKa = 4.02KK429 pKa = 9.77RR430 pKa = 11.84RR431 pKa = 11.84SGSEE435 pKa = 3.59YY436 pKa = 9.86RR437 pKa = 11.84RR438 pKa = 11.84GGAEE442 pKa = 3.63RR443 pKa = 11.84KK444 pKa = 9.85RR445 pKa = 11.84IAEE448 pKa = 4.0LKK450 pKa = 9.8EE451 pKa = 4.31RR452 pKa = 11.84EE453 pKa = 4.23IKK455 pKa = 10.48PDD457 pKa = 2.95VTAIMDD463 pKa = 4.04NLPGPKK469 pKa = 9.19HH470 pKa = 6.54SPINRR475 pKa = 11.84PNRR478 pKa = 11.84QKK480 pKa = 10.57QVEE483 pKa = 4.56FVTT486 pKa = 4.2
MM1 pKa = 7.44GFLRR5 pKa = 11.84RR6 pKa = 11.84VAGVSLRR13 pKa = 11.84DD14 pKa = 3.38RR15 pKa = 11.84RR16 pKa = 11.84PQQRR20 pKa = 11.84RR21 pKa = 11.84KK22 pKa = 9.82QKK24 pKa = 10.56PNTPSTKK31 pKa = 10.11RR32 pKa = 11.84LTSQHH37 pKa = 5.78SAQGTSEE44 pKa = 4.44SKK46 pKa = 10.7AHH48 pKa = 6.44PLSPAEE54 pKa = 4.96DD55 pKa = 3.3KK56 pKa = 10.79MEE58 pKa = 4.86PKK60 pKa = 9.97VQPDD64 pKa = 3.82TQPCSHH70 pKa = 7.13RR71 pKa = 11.84EE72 pKa = 3.86HH73 pKa = 7.07KK74 pKa = 9.14PTGFRR79 pKa = 11.84VSRR82 pKa = 11.84HH83 pKa = 4.12TAAFSCHH90 pKa = 6.27RR91 pKa = 11.84LSDD94 pKa = 3.83SSPTLQEE101 pKa = 3.99SLSPKK106 pKa = 10.52AEE108 pKa = 3.94VGGLAGHH115 pKa = 6.82GEE117 pKa = 4.3GDD119 pKa = 3.45SDD121 pKa = 3.97TDD123 pKa = 3.6LSEE126 pKa = 4.44SEE128 pKa = 4.6RR129 pKa = 11.84LPVSPSGGVPPQLQLRR145 pKa = 11.84PEE147 pKa = 4.38VIEE150 pKa = 5.39AEE152 pKa = 4.31HH153 pKa = 6.55CPSRR157 pKa = 11.84SHH159 pKa = 7.25RR160 pKa = 11.84PRR162 pKa = 11.84GHH164 pKa = 5.57NHH166 pKa = 6.04GAFDD170 pKa = 4.99FPDD173 pKa = 4.44FLPPPFNSWSLSQLAVFYY191 pKa = 11.14NMEE194 pKa = 4.17GRR196 pKa = 11.84GGPRR200 pKa = 11.84PRR202 pKa = 11.84PMGPLEE208 pKa = 4.54RR209 pKa = 11.84YY210 pKa = 9.59LDD212 pKa = 3.76RR213 pKa = 11.84LLQLEE218 pKa = 4.24WHH220 pKa = 6.59QIQTVQGEE228 pKa = 4.66GRR230 pKa = 11.84KK231 pKa = 10.24SMGSDD236 pKa = 3.23VLSSCHH242 pKa = 6.44RR243 pKa = 11.84SHH245 pKa = 7.17AAASSRR251 pKa = 11.84LSSPKK256 pKa = 10.31CILQCQRR263 pKa = 11.84AFSLTFLSCLASHH276 pKa = 6.78SALLSGCACTLCRR289 pKa = 11.84IRR291 pKa = 11.84YY292 pKa = 5.9STCSTLCCRR301 pKa = 11.84SSHH304 pKa = 5.75HH305 pKa = 5.63THH307 pKa = 6.9HH308 pKa = 6.74SRR310 pKa = 11.84LSPMLEE316 pKa = 3.57RR317 pKa = 11.84RR318 pKa = 11.84GPTLLPKK325 pKa = 10.34RR326 pKa = 11.84SYY328 pKa = 11.11SEE330 pKa = 4.16NRR332 pKa = 11.84VKK334 pKa = 10.96SSDD337 pKa = 3.33RR338 pKa = 11.84SSAPRR343 pKa = 11.84SQVFSSPVRR352 pKa = 11.84TNSHH356 pKa = 6.07LRR358 pKa = 11.84RR359 pKa = 11.84MQASGNIRR367 pKa = 11.84NPVKK371 pKa = 10.49VANTKK376 pKa = 9.13PHH378 pKa = 4.9STARR382 pKa = 11.84GGSVGAGTDD391 pKa = 3.1RR392 pKa = 11.84VGALGDD398 pKa = 3.63VVDD401 pKa = 4.1YY402 pKa = 8.86RR403 pKa = 11.84TGGFRR408 pKa = 11.84RR409 pKa = 11.84RR410 pKa = 11.84SGSEE414 pKa = 3.34QRR416 pKa = 11.84RR417 pKa = 11.84GGVEE421 pKa = 3.85RR422 pKa = 11.84QQGASEE428 pKa = 4.02KK429 pKa = 9.77RR430 pKa = 11.84RR431 pKa = 11.84SGSEE435 pKa = 3.59YY436 pKa = 9.86RR437 pKa = 11.84RR438 pKa = 11.84GGAEE442 pKa = 3.63RR443 pKa = 11.84KK444 pKa = 9.85RR445 pKa = 11.84IAEE448 pKa = 4.0LKK450 pKa = 9.8EE451 pKa = 4.31RR452 pKa = 11.84EE453 pKa = 4.23IKK455 pKa = 10.48PDD457 pKa = 2.95VTAIMDD463 pKa = 4.04NLPGPKK469 pKa = 9.19HH470 pKa = 6.54SPINRR475 pKa = 11.84PNRR478 pKa = 11.84QKK480 pKa = 10.57QVEE483 pKa = 4.56FVTT486 pKa = 4.2
Molecular weight: 53.8 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
12322852 |
50 |
22184 |
569.3 |
63.43 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.692 ± 0.015 | 2.209 ± 0.015 |
5.232 ± 0.011 | 6.965 ± 0.023 |
3.461 ± 0.014 | 6.452 ± 0.022 |
2.628 ± 0.009 | 4.24 ± 0.013 |
5.578 ± 0.02 | 9.463 ± 0.023 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.392 ± 0.007 | 3.79 ± 0.01 |
5.808 ± 0.022 | 4.825 ± 0.02 |
5.697 ± 0.014 | 8.775 ± 0.024 |
5.664 ± 0.013 | 6.318 ± 0.013 |
1.141 ± 0.006 | 2.67 ± 0.01 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
