
Maize chlorotic mottle virus (isolate United States/Kansas/1987) (MCMV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Tolucaviricetes; Tolivirales; Tombusviridae; Procedovirinae; Machlomovirus; Maize chlorotic mottle virus
Average proteome isoelectric point is 6.98
Get precalculated fractions of proteins
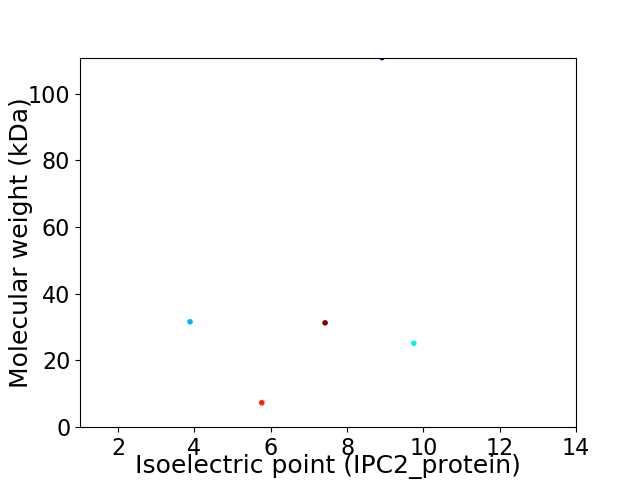
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P11640|RDRP_MCMVK RNA-directed RNA polymerase OS=Maize chlorotic mottle virus (isolate United States/Kansas/1987) OX=882210 GN=ORF2 PE=3 SV=3
MM1 pKa = 8.05PSPCTYY7 pKa = 11.18GDD9 pKa = 3.25PTFNSRR15 pKa = 11.84ILEE18 pKa = 4.1AVVADD23 pKa = 3.77VLGGTEE29 pKa = 4.53DD30 pKa = 3.79DD31 pKa = 4.56GGPSLEE37 pKa = 4.3EE38 pKa = 3.71WFDD41 pKa = 3.64AQTLSDD47 pKa = 4.07YY48 pKa = 8.99TNCATDD54 pKa = 3.58PPMATVHH61 pKa = 5.35TRR63 pKa = 11.84EE64 pKa = 4.08NDD66 pKa = 3.14IKK68 pKa = 11.02SWTEE72 pKa = 3.4LSEE75 pKa = 4.08NFPDD79 pKa = 3.92LVRR82 pKa = 11.84YY83 pKa = 9.24PEE85 pKa = 4.28SLVEE89 pKa = 4.02TLLDD93 pKa = 3.71TEE95 pKa = 4.91HH96 pKa = 6.99EE97 pKa = 4.53CGHH100 pKa = 6.58FYY102 pKa = 10.79DD103 pKa = 5.49APDD106 pKa = 3.42SFQISVTAMFTDD118 pKa = 3.6RR119 pKa = 11.84CKK121 pKa = 11.04CQFCDD126 pKa = 3.65PNFQARR132 pKa = 11.84SLSRR136 pKa = 11.84ALLGPLPEE144 pKa = 6.34SGDD147 pKa = 3.49DD148 pKa = 4.08AEE150 pKa = 4.58WMEE153 pKa = 4.72QAYY156 pKa = 8.22TPDD159 pKa = 3.61AEE161 pKa = 4.76LFVNEE166 pKa = 4.67PTDD169 pKa = 4.38DD170 pKa = 5.42PIPTTDD176 pKa = 4.04CKK178 pKa = 10.94RR179 pKa = 11.84PIQPTWSVDD188 pKa = 3.6VYY190 pKa = 11.15SKK192 pKa = 10.84QVDD195 pKa = 4.09SDD197 pKa = 3.37WGLSDD202 pKa = 3.37STNVTVCSASSSLLPAGRR220 pKa = 11.84GGIYY224 pKa = 9.73FMCPRR229 pKa = 11.84PEE231 pKa = 5.09GIEE234 pKa = 4.06RR235 pKa = 11.84VCLQVGAKK243 pKa = 8.97IRR245 pKa = 11.84SGAPNSGTITNLPGGTGYY263 pKa = 8.34GTEE266 pKa = 4.05WSDD269 pKa = 5.29DD270 pKa = 3.78GYY272 pKa = 9.67EE273 pKa = 4.35TQWSDD278 pKa = 3.29GPYY281 pKa = 9.8SIPSGLSDD289 pKa = 3.3
MM1 pKa = 8.05PSPCTYY7 pKa = 11.18GDD9 pKa = 3.25PTFNSRR15 pKa = 11.84ILEE18 pKa = 4.1AVVADD23 pKa = 3.77VLGGTEE29 pKa = 4.53DD30 pKa = 3.79DD31 pKa = 4.56GGPSLEE37 pKa = 4.3EE38 pKa = 3.71WFDD41 pKa = 3.64AQTLSDD47 pKa = 4.07YY48 pKa = 8.99TNCATDD54 pKa = 3.58PPMATVHH61 pKa = 5.35TRR63 pKa = 11.84EE64 pKa = 4.08NDD66 pKa = 3.14IKK68 pKa = 11.02SWTEE72 pKa = 3.4LSEE75 pKa = 4.08NFPDD79 pKa = 3.92LVRR82 pKa = 11.84YY83 pKa = 9.24PEE85 pKa = 4.28SLVEE89 pKa = 4.02TLLDD93 pKa = 3.71TEE95 pKa = 4.91HH96 pKa = 6.99EE97 pKa = 4.53CGHH100 pKa = 6.58FYY102 pKa = 10.79DD103 pKa = 5.49APDD106 pKa = 3.42SFQISVTAMFTDD118 pKa = 3.6RR119 pKa = 11.84CKK121 pKa = 11.04CQFCDD126 pKa = 3.65PNFQARR132 pKa = 11.84SLSRR136 pKa = 11.84ALLGPLPEE144 pKa = 6.34SGDD147 pKa = 3.49DD148 pKa = 4.08AEE150 pKa = 4.58WMEE153 pKa = 4.72QAYY156 pKa = 8.22TPDD159 pKa = 3.61AEE161 pKa = 4.76LFVNEE166 pKa = 4.67PTDD169 pKa = 4.38DD170 pKa = 5.42PIPTTDD176 pKa = 4.04CKK178 pKa = 10.94RR179 pKa = 11.84PIQPTWSVDD188 pKa = 3.6VYY190 pKa = 11.15SKK192 pKa = 10.84QVDD195 pKa = 4.09SDD197 pKa = 3.37WGLSDD202 pKa = 3.37STNVTVCSASSSLLPAGRR220 pKa = 11.84GGIYY224 pKa = 9.73FMCPRR229 pKa = 11.84PEE231 pKa = 5.09GIEE234 pKa = 4.06RR235 pKa = 11.84VCLQVGAKK243 pKa = 8.97IRR245 pKa = 11.84SGAPNSGTITNLPGGTGYY263 pKa = 8.34GTEE266 pKa = 4.05WSDD269 pKa = 5.29DD270 pKa = 3.78GYY272 pKa = 9.67EE273 pKa = 4.35TQWSDD278 pKa = 3.29GPYY281 pKa = 9.8SIPSGLSDD289 pKa = 3.3
Molecular weight: 31.59 kDa
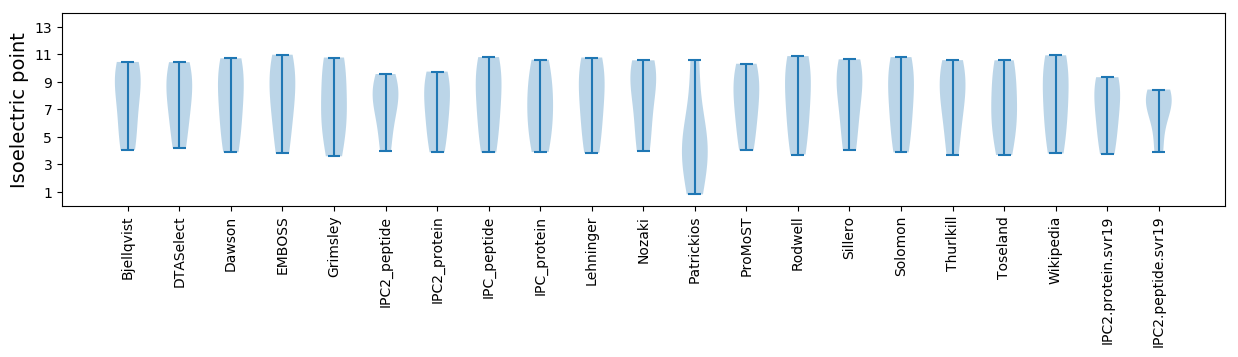
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P11642|CAPSD_MCMVK Capsid protein OS=Maize chlorotic mottle virus (isolate United States/Kansas/1987) OX=882210 GN=ORF4 PE=3 SV=1
MM1 pKa = 7.39AASSRR6 pKa = 11.84STRR9 pKa = 11.84GRR11 pKa = 11.84KK12 pKa = 7.81QRR14 pKa = 11.84GRR16 pKa = 11.84SVEE19 pKa = 4.0AKK21 pKa = 10.19SRR23 pKa = 11.84AIRR26 pKa = 11.84ANPPVPRR33 pKa = 11.84PNPQRR38 pKa = 11.84NRR40 pKa = 11.84PPPAGTTCSMSEE52 pKa = 3.57ILLAVSATTADD63 pKa = 3.85QILEE67 pKa = 4.13IPVCAGIDD75 pKa = 3.95FPAGTSPRR83 pKa = 11.84YY84 pKa = 9.62IGAAKK89 pKa = 9.45WLAAQSQMWNTIVFNSVRR107 pKa = 11.84ITWEE111 pKa = 3.65TFTADD116 pKa = 3.25TTSGYY121 pKa = 10.62ISMAFLSDD129 pKa = 3.68YY130 pKa = 9.81MLSIPTGVEE139 pKa = 3.6DD140 pKa = 3.57VARR143 pKa = 11.84IVPSATIALKK153 pKa = 10.74NRR155 pKa = 11.84GPSIVMPQNRR165 pKa = 11.84TVFRR169 pKa = 11.84CIQAGQFAALGSAADD184 pKa = 3.64KK185 pKa = 11.04QMYY188 pKa = 9.84SPGRR192 pKa = 11.84FIVAIPKK199 pKa = 10.33ASATQAVGQIKK210 pKa = 9.98ISYY213 pKa = 7.72SVSYY217 pKa = 10.16RR218 pKa = 11.84GAAILQPALVPGPGLANHH236 pKa = 7.38
MM1 pKa = 7.39AASSRR6 pKa = 11.84STRR9 pKa = 11.84GRR11 pKa = 11.84KK12 pKa = 7.81QRR14 pKa = 11.84GRR16 pKa = 11.84SVEE19 pKa = 4.0AKK21 pKa = 10.19SRR23 pKa = 11.84AIRR26 pKa = 11.84ANPPVPRR33 pKa = 11.84PNPQRR38 pKa = 11.84NRR40 pKa = 11.84PPPAGTTCSMSEE52 pKa = 3.57ILLAVSATTADD63 pKa = 3.85QILEE67 pKa = 4.13IPVCAGIDD75 pKa = 3.95FPAGTSPRR83 pKa = 11.84YY84 pKa = 9.62IGAAKK89 pKa = 9.45WLAAQSQMWNTIVFNSVRR107 pKa = 11.84ITWEE111 pKa = 3.65TFTADD116 pKa = 3.25TTSGYY121 pKa = 10.62ISMAFLSDD129 pKa = 3.68YY130 pKa = 9.81MLSIPTGVEE139 pKa = 3.6DD140 pKa = 3.57VARR143 pKa = 11.84IVPSATIALKK153 pKa = 10.74NRR155 pKa = 11.84GPSIVMPQNRR165 pKa = 11.84TVFRR169 pKa = 11.84CIQAGQFAALGSAADD184 pKa = 3.64KK185 pKa = 11.04QMYY188 pKa = 9.84SPGRR192 pKa = 11.84FIVAIPKK199 pKa = 10.33ASATQAVGQIKK210 pKa = 9.98ISYY213 pKa = 7.72SVSYY217 pKa = 10.16RR218 pKa = 11.84GAAILQPALVPGPGLANHH236 pKa = 7.38
Molecular weight: 25.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1833 |
64 |
965 |
366.6 |
41.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.819 ± 1.321 | 2.4 ± 0.489 |
4.637 ± 1.049 | 5.346 ± 0.751 |
3.873 ± 0.319 | 6.219 ± 0.413 |
2.182 ± 0.486 | 5.074 ± 0.846 |
3.983 ± 0.701 | 8.62 ± 0.999 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.291 ± 0.381 | 3.819 ± 0.22 |
6.601 ± 0.76 | 5.128 ± 0.822 |
7.147 ± 0.873 | 7.529 ± 0.937 |
6.874 ± 0.359 | 6.274 ± 0.227 |
2.073 ± 0.171 | 3.001 ± 0.514 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
