
Salinarchaeum sp. Harcht-Bsk1
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Natrialbales; Natrialbaceae; Salinarchaeum; unclassified Salinarchaeum
Average proteome isoelectric point is 4.7
Get precalculated fractions of proteins
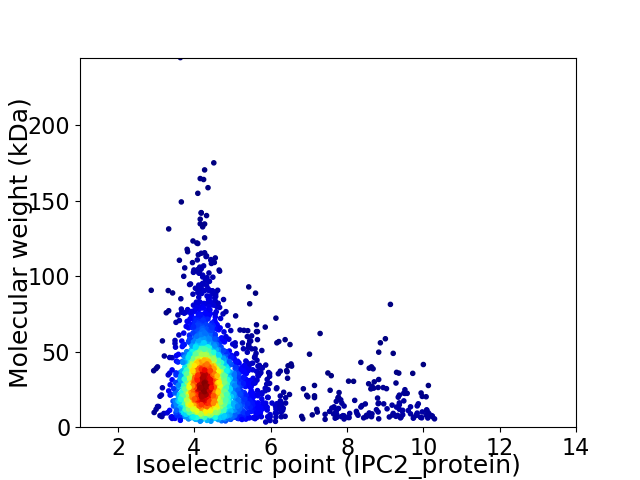
Virtual 2D-PAGE plot for 3013 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R4VYG8|R4VYG8_9EURY Peptide methionine sulfoxide reductase MsrB OS=Salinarchaeum sp. Harcht-Bsk1 OX=1333523 GN=msrB PE=3 SV=1
MM1 pKa = 7.74TDD3 pKa = 3.66DD4 pKa = 4.18DD5 pKa = 4.33TTIEE9 pKa = 3.98LSRR12 pKa = 11.84RR13 pKa = 11.84TVLGGLGGIGVAATGAALGTSAYY36 pKa = 8.83FTDD39 pKa = 5.09DD40 pKa = 3.06EE41 pKa = 4.6AFEE44 pKa = 4.71GNSLAAGSLDD54 pKa = 5.25LKK56 pKa = 11.03VDD58 pKa = 3.07WQEE61 pKa = 3.8WYY63 pKa = 10.87NGTKK67 pKa = 9.62IEE69 pKa = 5.28AYY71 pKa = 9.21PDD73 pKa = 4.0PDD75 pKa = 3.47NDD77 pKa = 3.8GVQDD81 pKa = 4.06AFASEE86 pKa = 4.45PGQTTAKK93 pKa = 10.12GVGYY97 pKa = 10.2VCEE100 pKa = 5.34DD101 pKa = 3.83GADD104 pKa = 3.77TPADD108 pKa = 4.13LDD110 pKa = 4.14PEE112 pKa = 4.5HH113 pKa = 7.0SLRR116 pKa = 11.84TTVSGEE122 pKa = 4.25TFGSGPPLPLVHH134 pKa = 6.66LTDD137 pKa = 5.42LKK139 pKa = 11.15PGDD142 pKa = 4.39CGGLTLSFHH151 pKa = 6.87LCDD154 pKa = 3.42NPGFVWMNGTLTEE167 pKa = 4.02NAEE170 pKa = 4.14NDD172 pKa = 3.43VTEE175 pKa = 4.74PEE177 pKa = 4.4SAVDD181 pKa = 4.22DD182 pKa = 3.92SDD184 pKa = 4.52GQGEE188 pKa = 4.18LADD191 pKa = 5.1AIRR194 pKa = 11.84TRR196 pKa = 11.84LWYY199 pKa = 10.0DD200 pKa = 3.53ADD202 pKa = 4.26CDD204 pKa = 4.04GEE206 pKa = 4.57RR207 pKa = 11.84DD208 pKa = 3.88DD209 pKa = 5.71GEE211 pKa = 4.52VPITPEE217 pKa = 3.63LSLAEE222 pKa = 3.98LLGVLGSDD230 pKa = 4.13HH231 pKa = 7.51GIPLDD236 pKa = 4.13GALGDD241 pKa = 4.64DD242 pKa = 4.32GATIAQAPSLGPGTCPEE259 pKa = 4.09TPYY262 pKa = 11.47DD263 pKa = 3.78EE264 pKa = 4.49VTTGTEE270 pKa = 3.88EE271 pKa = 4.32FPNGTTRR278 pKa = 11.84QRR280 pKa = 11.84MNPKK284 pKa = 10.0CSDD287 pKa = 3.44FGLVEE292 pKa = 4.12AVRR295 pKa = 11.84IGSDD299 pKa = 3.51EE300 pKa = 4.34PGGPLPAPGACATYY314 pKa = 8.42EE315 pKa = 3.88TDD317 pKa = 3.47YY318 pKa = 11.71GDD320 pKa = 4.07VRR322 pKa = 11.84VCRR325 pKa = 11.84DD326 pKa = 3.08ADD328 pKa = 3.64GTITSWEE335 pKa = 4.24TDD337 pKa = 3.52DD338 pKa = 5.87SPDD341 pKa = 3.41GSFDD345 pKa = 3.38ASGDD349 pKa = 3.75GFCVSKK355 pKa = 11.11VVVKK359 pKa = 10.76GGNQGANIYY368 pKa = 10.48HH369 pKa = 6.98YY370 pKa = 10.92DD371 pKa = 3.57RR372 pKa = 11.84DD373 pKa = 3.81ADD375 pKa = 3.76PVEE378 pKa = 4.5RR379 pKa = 11.84SSGDD383 pKa = 3.01EE384 pKa = 4.06GSFVTPTGQDD394 pKa = 3.37YY395 pKa = 11.36SHH397 pKa = 7.22VSVCVDD403 pKa = 3.99LVAEE407 pKa = 5.2DD408 pKa = 4.46GNGDD412 pKa = 3.72DD413 pKa = 6.11GRR415 pKa = 11.84DD416 pKa = 3.48CFEE419 pKa = 4.91NSTTHH424 pKa = 6.68CVGLEE429 pKa = 3.2WWLPVDD435 pKa = 5.2RR436 pKa = 11.84GNEE439 pKa = 3.92LQTDD443 pKa = 3.9SVGFDD448 pKa = 2.76VGFYY452 pKa = 10.99AEE454 pKa = 4.14QCRR457 pKa = 11.84HH458 pKa = 6.11NDD460 pKa = 4.74DD461 pKa = 4.09PPQTQRR467 pKa = 11.84IEE469 pKa = 4.12LGKK472 pKa = 10.37DD473 pKa = 2.76AARR476 pKa = 11.84NTGFEE481 pKa = 4.14AVSDD485 pKa = 3.62GGYY488 pKa = 10.23FGSGYY493 pKa = 10.09WKK495 pKa = 10.27DD496 pKa = 5.44DD497 pKa = 3.52PTQTNQDD504 pKa = 3.19GGTYY508 pKa = 8.57EE509 pKa = 3.97QLYY512 pKa = 8.4LTFDD516 pKa = 3.45QSFGPYY522 pKa = 8.74YY523 pKa = 10.81VSALAPFTIGDD534 pKa = 3.79IQSIRR539 pKa = 11.84YY540 pKa = 6.81RR541 pKa = 11.84TNTPSGVSQNYY552 pKa = 8.88FLEE555 pKa = 5.46IYY557 pKa = 8.9TSPDD561 pKa = 2.78GSNDD565 pKa = 3.26DD566 pKa = 3.74ASWYY570 pKa = 10.38GRR572 pKa = 11.84LLQALPGDD580 pKa = 3.87ALNRR584 pKa = 11.84TVSPDD589 pKa = 2.59TWVTWRR595 pKa = 11.84TEE597 pKa = 3.71SGTNQLTFYY606 pKa = 10.76DD607 pKa = 4.46HH608 pKa = 6.57NHH610 pKa = 7.35DD611 pKa = 4.1YY612 pKa = 9.55DD613 pKa = 4.26TTDD616 pKa = 3.38STPGDD621 pKa = 3.77APNAYY626 pKa = 9.77LGQDD630 pKa = 3.43TGVTLADD637 pKa = 4.2LQSTDD642 pKa = 3.14AFDD645 pKa = 3.27WSNYY649 pKa = 9.0VSDD652 pKa = 5.28ADD654 pKa = 4.02STPKK658 pKa = 10.42NYY660 pKa = 9.55RR661 pKa = 11.84DD662 pKa = 3.48EE663 pKa = 4.2EE664 pKa = 4.2VRR666 pKa = 11.84ALRR669 pKa = 11.84FATGSAWEE677 pKa = 4.95DD678 pKa = 3.38DD679 pKa = 4.34HH680 pKa = 7.25EE681 pKa = 5.31GCLDD685 pKa = 4.27AIEE688 pKa = 4.42ITLEE692 pKa = 4.17DD693 pKa = 3.46GRR695 pKa = 11.84SAVVDD700 pKa = 4.2LEE702 pKa = 4.24PP703 pKa = 5.71
MM1 pKa = 7.74TDD3 pKa = 3.66DD4 pKa = 4.18DD5 pKa = 4.33TTIEE9 pKa = 3.98LSRR12 pKa = 11.84RR13 pKa = 11.84TVLGGLGGIGVAATGAALGTSAYY36 pKa = 8.83FTDD39 pKa = 5.09DD40 pKa = 3.06EE41 pKa = 4.6AFEE44 pKa = 4.71GNSLAAGSLDD54 pKa = 5.25LKK56 pKa = 11.03VDD58 pKa = 3.07WQEE61 pKa = 3.8WYY63 pKa = 10.87NGTKK67 pKa = 9.62IEE69 pKa = 5.28AYY71 pKa = 9.21PDD73 pKa = 4.0PDD75 pKa = 3.47NDD77 pKa = 3.8GVQDD81 pKa = 4.06AFASEE86 pKa = 4.45PGQTTAKK93 pKa = 10.12GVGYY97 pKa = 10.2VCEE100 pKa = 5.34DD101 pKa = 3.83GADD104 pKa = 3.77TPADD108 pKa = 4.13LDD110 pKa = 4.14PEE112 pKa = 4.5HH113 pKa = 7.0SLRR116 pKa = 11.84TTVSGEE122 pKa = 4.25TFGSGPPLPLVHH134 pKa = 6.66LTDD137 pKa = 5.42LKK139 pKa = 11.15PGDD142 pKa = 4.39CGGLTLSFHH151 pKa = 6.87LCDD154 pKa = 3.42NPGFVWMNGTLTEE167 pKa = 4.02NAEE170 pKa = 4.14NDD172 pKa = 3.43VTEE175 pKa = 4.74PEE177 pKa = 4.4SAVDD181 pKa = 4.22DD182 pKa = 3.92SDD184 pKa = 4.52GQGEE188 pKa = 4.18LADD191 pKa = 5.1AIRR194 pKa = 11.84TRR196 pKa = 11.84LWYY199 pKa = 10.0DD200 pKa = 3.53ADD202 pKa = 4.26CDD204 pKa = 4.04GEE206 pKa = 4.57RR207 pKa = 11.84DD208 pKa = 3.88DD209 pKa = 5.71GEE211 pKa = 4.52VPITPEE217 pKa = 3.63LSLAEE222 pKa = 3.98LLGVLGSDD230 pKa = 4.13HH231 pKa = 7.51GIPLDD236 pKa = 4.13GALGDD241 pKa = 4.64DD242 pKa = 4.32GATIAQAPSLGPGTCPEE259 pKa = 4.09TPYY262 pKa = 11.47DD263 pKa = 3.78EE264 pKa = 4.49VTTGTEE270 pKa = 3.88EE271 pKa = 4.32FPNGTTRR278 pKa = 11.84QRR280 pKa = 11.84MNPKK284 pKa = 10.0CSDD287 pKa = 3.44FGLVEE292 pKa = 4.12AVRR295 pKa = 11.84IGSDD299 pKa = 3.51EE300 pKa = 4.34PGGPLPAPGACATYY314 pKa = 8.42EE315 pKa = 3.88TDD317 pKa = 3.47YY318 pKa = 11.71GDD320 pKa = 4.07VRR322 pKa = 11.84VCRR325 pKa = 11.84DD326 pKa = 3.08ADD328 pKa = 3.64GTITSWEE335 pKa = 4.24TDD337 pKa = 3.52DD338 pKa = 5.87SPDD341 pKa = 3.41GSFDD345 pKa = 3.38ASGDD349 pKa = 3.75GFCVSKK355 pKa = 11.11VVVKK359 pKa = 10.76GGNQGANIYY368 pKa = 10.48HH369 pKa = 6.98YY370 pKa = 10.92DD371 pKa = 3.57RR372 pKa = 11.84DD373 pKa = 3.81ADD375 pKa = 3.76PVEE378 pKa = 4.5RR379 pKa = 11.84SSGDD383 pKa = 3.01EE384 pKa = 4.06GSFVTPTGQDD394 pKa = 3.37YY395 pKa = 11.36SHH397 pKa = 7.22VSVCVDD403 pKa = 3.99LVAEE407 pKa = 5.2DD408 pKa = 4.46GNGDD412 pKa = 3.72DD413 pKa = 6.11GRR415 pKa = 11.84DD416 pKa = 3.48CFEE419 pKa = 4.91NSTTHH424 pKa = 6.68CVGLEE429 pKa = 3.2WWLPVDD435 pKa = 5.2RR436 pKa = 11.84GNEE439 pKa = 3.92LQTDD443 pKa = 3.9SVGFDD448 pKa = 2.76VGFYY452 pKa = 10.99AEE454 pKa = 4.14QCRR457 pKa = 11.84HH458 pKa = 6.11NDD460 pKa = 4.74DD461 pKa = 4.09PPQTQRR467 pKa = 11.84IEE469 pKa = 4.12LGKK472 pKa = 10.37DD473 pKa = 2.76AARR476 pKa = 11.84NTGFEE481 pKa = 4.14AVSDD485 pKa = 3.62GGYY488 pKa = 10.23FGSGYY493 pKa = 10.09WKK495 pKa = 10.27DD496 pKa = 5.44DD497 pKa = 3.52PTQTNQDD504 pKa = 3.19GGTYY508 pKa = 8.57EE509 pKa = 3.97QLYY512 pKa = 8.4LTFDD516 pKa = 3.45QSFGPYY522 pKa = 8.74YY523 pKa = 10.81VSALAPFTIGDD534 pKa = 3.79IQSIRR539 pKa = 11.84YY540 pKa = 6.81RR541 pKa = 11.84TNTPSGVSQNYY552 pKa = 8.88FLEE555 pKa = 5.46IYY557 pKa = 8.9TSPDD561 pKa = 2.78GSNDD565 pKa = 3.26DD566 pKa = 3.74ASWYY570 pKa = 10.38GRR572 pKa = 11.84LLQALPGDD580 pKa = 3.87ALNRR584 pKa = 11.84TVSPDD589 pKa = 2.59TWVTWRR595 pKa = 11.84TEE597 pKa = 3.71SGTNQLTFYY606 pKa = 10.76DD607 pKa = 4.46HH608 pKa = 6.57NHH610 pKa = 7.35DD611 pKa = 4.1YY612 pKa = 9.55DD613 pKa = 4.26TTDD616 pKa = 3.38STPGDD621 pKa = 3.77APNAYY626 pKa = 9.77LGQDD630 pKa = 3.43TGVTLADD637 pKa = 4.2LQSTDD642 pKa = 3.14AFDD645 pKa = 3.27WSNYY649 pKa = 9.0VSDD652 pKa = 5.28ADD654 pKa = 4.02STPKK658 pKa = 10.42NYY660 pKa = 9.55RR661 pKa = 11.84DD662 pKa = 3.48EE663 pKa = 4.2EE664 pKa = 4.2VRR666 pKa = 11.84ALRR669 pKa = 11.84FATGSAWEE677 pKa = 4.95DD678 pKa = 3.38DD679 pKa = 4.34HH680 pKa = 7.25EE681 pKa = 5.31GCLDD685 pKa = 4.27AIEE688 pKa = 4.42ITLEE692 pKa = 4.17DD693 pKa = 3.46GRR695 pKa = 11.84SAVVDD700 pKa = 4.2LEE702 pKa = 4.24PP703 pKa = 5.71
Molecular weight: 75.41 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R4W6K2|R4W6K2_9EURY UDP-glucose 4-epimerase OS=Salinarchaeum sp. Harcht-Bsk1 OX=1333523 GN=L593_02460 PE=4 SV=1
MM1 pKa = 7.44LFYY4 pKa = 11.02YY5 pKa = 9.99EE6 pKa = 4.82SQSQPISLKK15 pKa = 10.38KK16 pKa = 10.35DD17 pKa = 3.11DD18 pKa = 4.15FAQIVLMRR26 pKa = 11.84CGRR29 pKa = 11.84QSLVMEE35 pKa = 4.41VEE37 pKa = 3.97RR38 pKa = 11.84RR39 pKa = 11.84PLRR42 pKa = 11.84TSLTYY47 pKa = 11.01SLDD50 pKa = 3.31VRR52 pKa = 11.84CNTRR56 pKa = 11.84VFKK59 pKa = 10.85RR60 pKa = 11.84RR61 pKa = 11.84FSVPFIKK68 pKa = 10.29QRR70 pKa = 11.84IKK72 pKa = 9.71MVKK75 pKa = 9.93RR76 pKa = 11.84NN77 pKa = 3.37
MM1 pKa = 7.44LFYY4 pKa = 11.02YY5 pKa = 9.99EE6 pKa = 4.82SQSQPISLKK15 pKa = 10.38KK16 pKa = 10.35DD17 pKa = 3.11DD18 pKa = 4.15FAQIVLMRR26 pKa = 11.84CGRR29 pKa = 11.84QSLVMEE35 pKa = 4.41VEE37 pKa = 3.97RR38 pKa = 11.84RR39 pKa = 11.84PLRR42 pKa = 11.84TSLTYY47 pKa = 11.01SLDD50 pKa = 3.31VRR52 pKa = 11.84CNTRR56 pKa = 11.84VFKK59 pKa = 10.85RR60 pKa = 11.84RR61 pKa = 11.84FSVPFIKK68 pKa = 10.29QRR70 pKa = 11.84IKK72 pKa = 9.71MVKK75 pKa = 9.93RR76 pKa = 11.84NN77 pKa = 3.37
Molecular weight: 9.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
911803 |
31 |
2381 |
302.6 |
32.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.658 ± 0.067 | 0.686 ± 0.014 |
8.843 ± 0.052 | 8.881 ± 0.062 |
3.111 ± 0.029 | 8.833 ± 0.04 |
2.048 ± 0.021 | 4.013 ± 0.03 |
1.479 ± 0.024 | 8.794 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.637 ± 0.021 | 2.079 ± 0.027 |
4.815 ± 0.029 | 2.577 ± 0.027 |
6.355 ± 0.044 | 5.398 ± 0.041 |
6.154 ± 0.042 | 8.989 ± 0.043 |
1.14 ± 0.018 | 2.511 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
