
Indibacter alkaliphilus LW1
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Cytophagia; Cytophagales; Cyclobacteriaceae; Indibacter; Indibacter alkaliphilus
Average proteome isoelectric point is 6.36
Get precalculated fractions of proteins
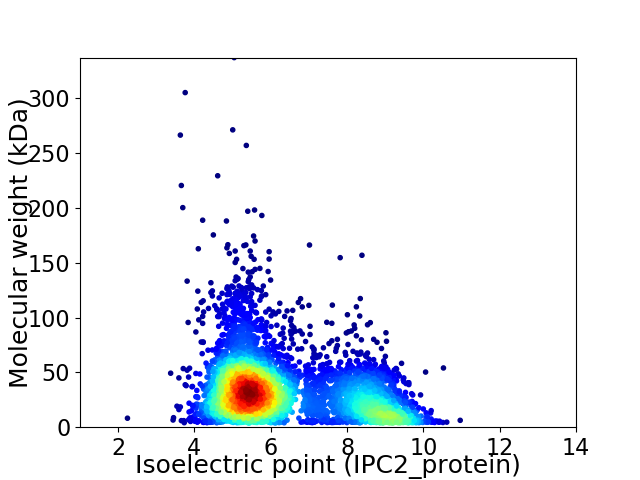
Virtual 2D-PAGE plot for 4650 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|S2DM90|S2DM90_9BACT Uncharacterized protein OS=Indibacter alkaliphilus LW1 OX=1189612 GN=A33Q_0106 PE=4 SV=1
MM1 pKa = 7.84RR2 pKa = 11.84KK3 pKa = 9.22LLKK6 pKa = 10.38YY7 pKa = 10.39LLVVLPFVVMAACVDD22 pKa = 3.66EE23 pKa = 4.9QNIEE27 pKa = 4.16DD28 pKa = 4.67LPPTSEE34 pKa = 4.31DD35 pKa = 2.91ATFSVEE41 pKa = 4.14PTSEE45 pKa = 3.81NPNIVNFTADD55 pKa = 3.26RR56 pKa = 11.84EE57 pKa = 4.32FFRR60 pKa = 11.84MQWDD64 pKa = 4.08LGNGTTVEE72 pKa = 4.1GRR74 pKa = 11.84TVTGTYY80 pKa = 9.82PNEE83 pKa = 3.72GTYY86 pKa = 10.03TVTLTVFNAAGSASFSQDD104 pKa = 2.32IVIDD108 pKa = 3.68QTDD111 pKa = 3.83PTLLDD116 pKa = 3.49SPIFNILTGGINAIEE131 pKa = 4.27GRR133 pKa = 11.84TWVIDD138 pKa = 3.6SARR141 pKa = 11.84VGHH144 pKa = 6.51FGVGPNPSQAGDD156 pKa = 3.56FPEE159 pKa = 4.8WYY161 pKa = 9.86AAQANEE167 pKa = 3.85KK168 pKa = 9.27RR169 pKa = 11.84GSGMYY174 pKa = 8.93TDD176 pKa = 5.11RR177 pKa = 11.84YY178 pKa = 9.18TFFLDD183 pKa = 3.25NFGFNMEE190 pKa = 4.53TNGFVYY196 pKa = 10.49LNEE199 pKa = 4.09AQGPNFPGAFDD210 pKa = 4.56PGVGDD215 pKa = 4.27LSAPYY220 pKa = 8.61EE221 pKa = 4.41APDD224 pKa = 3.72GLTWSYY230 pKa = 11.42TEE232 pKa = 4.09VEE234 pKa = 4.16NGFDD238 pKa = 3.56EE239 pKa = 5.13LRR241 pKa = 11.84ISEE244 pKa = 4.32GGFLGYY250 pKa = 9.96FAGGTTYY257 pKa = 10.63QIVNLEE263 pKa = 4.12EE264 pKa = 4.7NEE266 pKa = 4.03MFLRR270 pKa = 11.84YY271 pKa = 8.81IDD273 pKa = 3.64QANEE277 pKa = 3.33EE278 pKa = 4.32LAWYY282 pKa = 9.4IRR284 pKa = 11.84LIPAGFDD291 pKa = 3.12SGEE294 pKa = 4.08DD295 pKa = 3.59VQEE298 pKa = 4.36EE299 pKa = 4.52EE300 pKa = 4.43PSDD303 pKa = 3.63VNFNFEE309 pKa = 4.58DD310 pKa = 4.55LIGDD314 pKa = 4.47GTQAWTLKK322 pKa = 10.11SAAGAFGVGPAPGNDD337 pKa = 2.81SFFPNGNDD345 pKa = 3.0ISDD348 pKa = 3.64EE349 pKa = 4.13RR350 pKa = 11.84ACLFNDD356 pKa = 4.49LYY358 pKa = 10.77IFSEE362 pKa = 4.51DD363 pKa = 2.87GTYY366 pKa = 10.33EE367 pKa = 4.11YY368 pKa = 8.98DD369 pKa = 3.47TQGDD373 pKa = 3.77IFAEE377 pKa = 5.12FYY379 pKa = 10.35MGVSEE384 pKa = 5.44EE385 pKa = 4.48GCQDD389 pKa = 3.27EE390 pKa = 5.06SNLEE394 pKa = 4.12GTPGEE399 pKa = 3.96AWGSGVHH406 pKa = 5.96SFEE409 pKa = 4.12FTPGEE414 pKa = 4.23GSTRR418 pKa = 11.84PKK420 pKa = 9.51ITVTGTGAFVVLAKK434 pKa = 10.58AFNGGEE440 pKa = 4.13YY441 pKa = 10.3EE442 pKa = 4.2QGPPNEE448 pKa = 4.46NASVTYY454 pKa = 10.44DD455 pKa = 3.39VLDD458 pKa = 3.83YY459 pKa = 11.25DD460 pKa = 4.48PEE462 pKa = 4.65SKK464 pKa = 10.58EE465 pKa = 3.77LTLTIDD471 pKa = 3.13ITNDD475 pKa = 2.92GGVWWTFVLVPNEE488 pKa = 3.75
MM1 pKa = 7.84RR2 pKa = 11.84KK3 pKa = 9.22LLKK6 pKa = 10.38YY7 pKa = 10.39LLVVLPFVVMAACVDD22 pKa = 3.66EE23 pKa = 4.9QNIEE27 pKa = 4.16DD28 pKa = 4.67LPPTSEE34 pKa = 4.31DD35 pKa = 2.91ATFSVEE41 pKa = 4.14PTSEE45 pKa = 3.81NPNIVNFTADD55 pKa = 3.26RR56 pKa = 11.84EE57 pKa = 4.32FFRR60 pKa = 11.84MQWDD64 pKa = 4.08LGNGTTVEE72 pKa = 4.1GRR74 pKa = 11.84TVTGTYY80 pKa = 9.82PNEE83 pKa = 3.72GTYY86 pKa = 10.03TVTLTVFNAAGSASFSQDD104 pKa = 2.32IVIDD108 pKa = 3.68QTDD111 pKa = 3.83PTLLDD116 pKa = 3.49SPIFNILTGGINAIEE131 pKa = 4.27GRR133 pKa = 11.84TWVIDD138 pKa = 3.6SARR141 pKa = 11.84VGHH144 pKa = 6.51FGVGPNPSQAGDD156 pKa = 3.56FPEE159 pKa = 4.8WYY161 pKa = 9.86AAQANEE167 pKa = 3.85KK168 pKa = 9.27RR169 pKa = 11.84GSGMYY174 pKa = 8.93TDD176 pKa = 5.11RR177 pKa = 11.84YY178 pKa = 9.18TFFLDD183 pKa = 3.25NFGFNMEE190 pKa = 4.53TNGFVYY196 pKa = 10.49LNEE199 pKa = 4.09AQGPNFPGAFDD210 pKa = 4.56PGVGDD215 pKa = 4.27LSAPYY220 pKa = 8.61EE221 pKa = 4.41APDD224 pKa = 3.72GLTWSYY230 pKa = 11.42TEE232 pKa = 4.09VEE234 pKa = 4.16NGFDD238 pKa = 3.56EE239 pKa = 5.13LRR241 pKa = 11.84ISEE244 pKa = 4.32GGFLGYY250 pKa = 9.96FAGGTTYY257 pKa = 10.63QIVNLEE263 pKa = 4.12EE264 pKa = 4.7NEE266 pKa = 4.03MFLRR270 pKa = 11.84YY271 pKa = 8.81IDD273 pKa = 3.64QANEE277 pKa = 3.33EE278 pKa = 4.32LAWYY282 pKa = 9.4IRR284 pKa = 11.84LIPAGFDD291 pKa = 3.12SGEE294 pKa = 4.08DD295 pKa = 3.59VQEE298 pKa = 4.36EE299 pKa = 4.52EE300 pKa = 4.43PSDD303 pKa = 3.63VNFNFEE309 pKa = 4.58DD310 pKa = 4.55LIGDD314 pKa = 4.47GTQAWTLKK322 pKa = 10.11SAAGAFGVGPAPGNDD337 pKa = 2.81SFFPNGNDD345 pKa = 3.0ISDD348 pKa = 3.64EE349 pKa = 4.13RR350 pKa = 11.84ACLFNDD356 pKa = 4.49LYY358 pKa = 10.77IFSEE362 pKa = 4.51DD363 pKa = 2.87GTYY366 pKa = 10.33EE367 pKa = 4.11YY368 pKa = 8.98DD369 pKa = 3.47TQGDD373 pKa = 3.77IFAEE377 pKa = 5.12FYY379 pKa = 10.35MGVSEE384 pKa = 5.44EE385 pKa = 4.48GCQDD389 pKa = 3.27EE390 pKa = 5.06SNLEE394 pKa = 4.12GTPGEE399 pKa = 3.96AWGSGVHH406 pKa = 5.96SFEE409 pKa = 4.12FTPGEE414 pKa = 4.23GSTRR418 pKa = 11.84PKK420 pKa = 9.51ITVTGTGAFVVLAKK434 pKa = 10.58AFNGGEE440 pKa = 4.13YY441 pKa = 10.3EE442 pKa = 4.2QGPPNEE448 pKa = 4.46NASVTYY454 pKa = 10.44DD455 pKa = 3.39VLDD458 pKa = 3.83YY459 pKa = 11.25DD460 pKa = 4.48PEE462 pKa = 4.65SKK464 pKa = 10.58EE465 pKa = 3.77LTLTIDD471 pKa = 3.13ITNDD475 pKa = 2.92GGVWWTFVLVPNEE488 pKa = 3.75
Molecular weight: 53.49 kDa
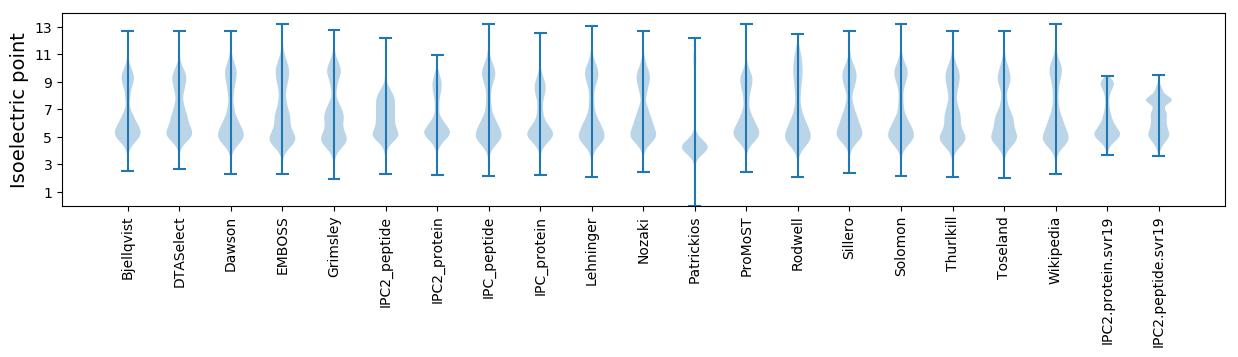
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|S2DKF0|S2DKF0_9BACT mRNA interferase OS=Indibacter alkaliphilus LW1 OX=1189612 GN=A33Q_0549 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84RR10 pKa = 11.84KK11 pKa = 9.62RR12 pKa = 11.84RR13 pKa = 11.84NKK15 pKa = 9.78HH16 pKa = 3.85GFRR19 pKa = 11.84LRR21 pKa = 11.84MSTANGRR28 pKa = 11.84RR29 pKa = 11.84VLKK32 pKa = 10.41SRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.19GRR39 pKa = 11.84HH40 pKa = 5.41KK41 pKa = 10.43LTVSSEE47 pKa = 4.01KK48 pKa = 9.88TLKK51 pKa = 10.51KK52 pKa = 10.72
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84RR10 pKa = 11.84KK11 pKa = 9.62RR12 pKa = 11.84RR13 pKa = 11.84NKK15 pKa = 9.78HH16 pKa = 3.85GFRR19 pKa = 11.84LRR21 pKa = 11.84MSTANGRR28 pKa = 11.84RR29 pKa = 11.84VLKK32 pKa = 10.41SRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.19GRR39 pKa = 11.84HH40 pKa = 5.41KK41 pKa = 10.43LTVSSEE47 pKa = 4.01KK48 pKa = 9.88TLKK51 pKa = 10.51KK52 pKa = 10.72
Molecular weight: 6.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1482599 |
37 |
2932 |
318.8 |
36.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.411 ± 0.032 | 0.659 ± 0.011 |
5.43 ± 0.029 | 7.236 ± 0.036 |
5.524 ± 0.031 | 7.04 ± 0.04 |
1.856 ± 0.017 | 7.31 ± 0.03 |
6.872 ± 0.05 | 9.757 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.461 ± 0.017 | 5.296 ± 0.034 |
3.872 ± 0.023 | 3.619 ± 0.021 |
4.149 ± 0.027 | 6.548 ± 0.028 |
4.781 ± 0.023 | 6.154 ± 0.031 |
1.255 ± 0.016 | 3.768 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
