
Togninia minima (strain UCR-PA7) (Esca disease fungus) (Phaeoacremonium aleophilum)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Sordariomycetes; Sordariomycetidae; Togniniales; Togniniaceae; Phaeoacremonium; Phaeoacremonium minimum
Average proteome isoelectric point is 6.16
Get precalculated fractions of proteins
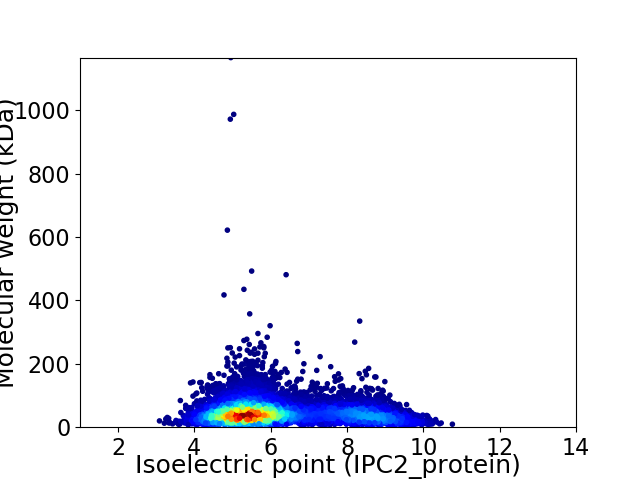
Virtual 2D-PAGE plot for 8833 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R8BHP3|R8BHP3_TOGMI Putative nad dependent epimerase dehydratase protein OS=Togninia minima (strain UCR-PA7) OX=1286976 GN=UCRPA7_5681 PE=4 SV=1
MM1 pKa = 7.0RR2 pKa = 11.84TVAAVAALLALAGSAIAGTLKK23 pKa = 10.55IPVHH27 pKa = 5.47KK28 pKa = 10.05QGPGSAVSKK37 pKa = 10.84HH38 pKa = 5.62PLLTKK43 pKa = 9.93RR44 pKa = 11.84ASAIEE49 pKa = 3.86EE50 pKa = 4.19LVNNITGGGYY60 pKa = 8.88YY61 pKa = 10.59ASVSVGTPGQSINLVLDD78 pKa = 3.76TGSSDD83 pKa = 3.49AFVVDD88 pKa = 4.89VNADD92 pKa = 3.31LCTSRR97 pKa = 11.84RR98 pKa = 11.84LQVEE102 pKa = 3.9YY103 pKa = 11.2GMTCDD108 pKa = 3.48ATFDD112 pKa = 4.04SSDD115 pKa = 3.07SSTFSVVQQNGFSIEE130 pKa = 4.06YY131 pKa = 9.87LDD133 pKa = 4.65GSTASGDD140 pKa = 3.73FVSDD144 pKa = 3.15SFEE147 pKa = 4.15IADD150 pKa = 3.59VTLDD154 pKa = 3.67DD155 pKa = 4.42FQFGLADD162 pKa = 3.57STVTGTGVLGIGFVAEE178 pKa = 4.29EE179 pKa = 4.44VALTTYY185 pKa = 10.83PNLPQALVDD194 pKa = 4.11AGAISVNAYY203 pKa = 10.16SLYY206 pKa = 10.44LNEE209 pKa = 4.48YY210 pKa = 10.57DD211 pKa = 4.77SDD213 pKa = 3.69TGSILFGGVDD223 pKa = 3.39TEE225 pKa = 4.24KK226 pKa = 10.92FIGEE230 pKa = 4.23LTVLPILQDD239 pKa = 3.23ARR241 pKa = 11.84GRR243 pKa = 11.84NYY245 pKa = 10.73SSFTIGLAEE254 pKa = 4.08LAAVFSNGTDD264 pKa = 3.47PANITLPSVLPAILDD279 pKa = 3.67SGTTLSYY286 pKa = 11.11LPDD289 pKa = 4.36DD290 pKa = 3.75VATPLFEE297 pKa = 4.76AVNAYY302 pKa = 8.91TYY304 pKa = 10.71TEE306 pKa = 4.68GSQGLTLIDD315 pKa = 4.19CKK317 pKa = 11.24YY318 pKa = 10.57LDD320 pKa = 3.99SDD322 pKa = 4.77EE323 pKa = 4.41EE324 pKa = 4.17LTMNFGFASSLTSSSSEE341 pKa = 3.62VTISVRR347 pKa = 11.84AAEE350 pKa = 4.47LVLDD354 pKa = 4.56VYY356 pKa = 10.86QGYY359 pKa = 9.4QSQMPSEE366 pKa = 4.19IPFDD370 pKa = 4.09EE371 pKa = 4.41VCLFGLQSSGVALPSGQAQSIEE393 pKa = 4.04FALLGDD399 pKa = 3.87TFLRR403 pKa = 11.84SAYY406 pKa = 9.88VVYY409 pKa = 10.43HH410 pKa = 6.06LEE412 pKa = 3.78NRR414 pKa = 11.84QIGLAQANLGSSDD427 pKa = 3.83SDD429 pKa = 3.78VQEE432 pKa = 4.56LSASGTSLPVFTGVASQASNPTATTTRR459 pKa = 11.84SSGGSGNGGGSSSTSQSGMVIITVTASPTASNNAAVASTRR499 pKa = 11.84PPTGEE504 pKa = 3.9AFAVMAVVGIFTLFGGLLFALL525 pKa = 4.79
MM1 pKa = 7.0RR2 pKa = 11.84TVAAVAALLALAGSAIAGTLKK23 pKa = 10.55IPVHH27 pKa = 5.47KK28 pKa = 10.05QGPGSAVSKK37 pKa = 10.84HH38 pKa = 5.62PLLTKK43 pKa = 9.93RR44 pKa = 11.84ASAIEE49 pKa = 3.86EE50 pKa = 4.19LVNNITGGGYY60 pKa = 8.88YY61 pKa = 10.59ASVSVGTPGQSINLVLDD78 pKa = 3.76TGSSDD83 pKa = 3.49AFVVDD88 pKa = 4.89VNADD92 pKa = 3.31LCTSRR97 pKa = 11.84RR98 pKa = 11.84LQVEE102 pKa = 3.9YY103 pKa = 11.2GMTCDD108 pKa = 3.48ATFDD112 pKa = 4.04SSDD115 pKa = 3.07SSTFSVVQQNGFSIEE130 pKa = 4.06YY131 pKa = 9.87LDD133 pKa = 4.65GSTASGDD140 pKa = 3.73FVSDD144 pKa = 3.15SFEE147 pKa = 4.15IADD150 pKa = 3.59VTLDD154 pKa = 3.67DD155 pKa = 4.42FQFGLADD162 pKa = 3.57STVTGTGVLGIGFVAEE178 pKa = 4.29EE179 pKa = 4.44VALTTYY185 pKa = 10.83PNLPQALVDD194 pKa = 4.11AGAISVNAYY203 pKa = 10.16SLYY206 pKa = 10.44LNEE209 pKa = 4.48YY210 pKa = 10.57DD211 pKa = 4.77SDD213 pKa = 3.69TGSILFGGVDD223 pKa = 3.39TEE225 pKa = 4.24KK226 pKa = 10.92FIGEE230 pKa = 4.23LTVLPILQDD239 pKa = 3.23ARR241 pKa = 11.84GRR243 pKa = 11.84NYY245 pKa = 10.73SSFTIGLAEE254 pKa = 4.08LAAVFSNGTDD264 pKa = 3.47PANITLPSVLPAILDD279 pKa = 3.67SGTTLSYY286 pKa = 11.11LPDD289 pKa = 4.36DD290 pKa = 3.75VATPLFEE297 pKa = 4.76AVNAYY302 pKa = 8.91TYY304 pKa = 10.71TEE306 pKa = 4.68GSQGLTLIDD315 pKa = 4.19CKK317 pKa = 11.24YY318 pKa = 10.57LDD320 pKa = 3.99SDD322 pKa = 4.77EE323 pKa = 4.41EE324 pKa = 4.17LTMNFGFASSLTSSSSEE341 pKa = 3.62VTISVRR347 pKa = 11.84AAEE350 pKa = 4.47LVLDD354 pKa = 4.56VYY356 pKa = 10.86QGYY359 pKa = 9.4QSQMPSEE366 pKa = 4.19IPFDD370 pKa = 4.09EE371 pKa = 4.41VCLFGLQSSGVALPSGQAQSIEE393 pKa = 4.04FALLGDD399 pKa = 3.87TFLRR403 pKa = 11.84SAYY406 pKa = 9.88VVYY409 pKa = 10.43HH410 pKa = 6.06LEE412 pKa = 3.78NRR414 pKa = 11.84QIGLAQANLGSSDD427 pKa = 3.83SDD429 pKa = 3.78VQEE432 pKa = 4.56LSASGTSLPVFTGVASQASNPTATTTRR459 pKa = 11.84SSGGSGNGGGSSSTSQSGMVIITVTASPTASNNAAVASTRR499 pKa = 11.84PPTGEE504 pKa = 3.9AFAVMAVVGIFTLFGGLLFALL525 pKa = 4.79
Molecular weight: 54.17 kDa
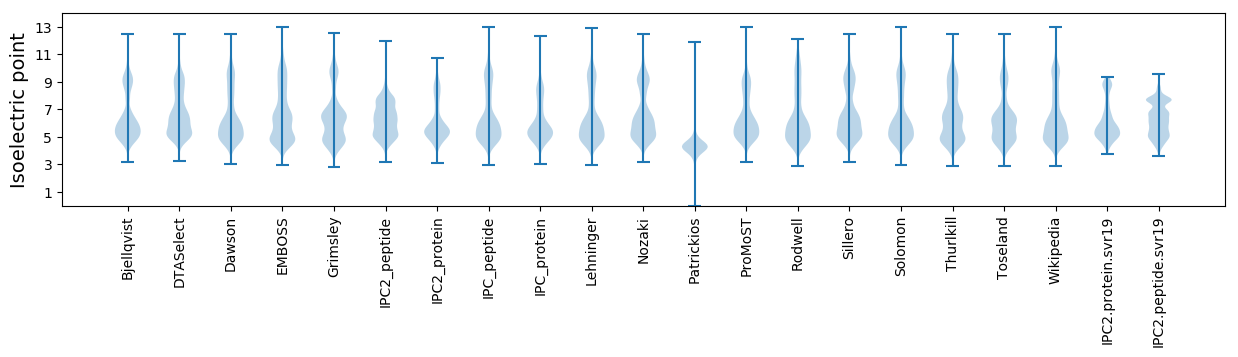
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R8BVY4|R8BVY4_TOGMI Ribosome biogenesis protein RLP24 OS=Togninia minima (strain UCR-PA7) OX=1286976 GN=UCRPA7_963 PE=3 SV=1
MM1 pKa = 6.93VGRR4 pKa = 11.84RR5 pKa = 11.84HH6 pKa = 5.89HH7 pKa = 6.51HH8 pKa = 5.09TTTTTTTRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84GLFSSRR24 pKa = 11.84RR25 pKa = 11.84QPVVVHH31 pKa = 5.1HH32 pKa = 6.06QKK34 pKa = 10.13RR35 pKa = 11.84HH36 pKa = 4.31ATLGDD41 pKa = 4.03KK42 pKa = 10.67ISGAFLKK49 pKa = 10.97LKK51 pKa = 10.56GSLTGRR57 pKa = 11.84PGEE60 pKa = 4.13KK61 pKa = 9.9AAGTRR66 pKa = 11.84RR67 pKa = 11.84MRR69 pKa = 11.84GTDD72 pKa = 2.74GRR74 pKa = 11.84GSHH77 pKa = 5.71RR78 pKa = 11.84RR79 pKa = 11.84RR80 pKa = 11.84RR81 pKa = 11.84FFF83 pKa = 4.33
MM1 pKa = 6.93VGRR4 pKa = 11.84RR5 pKa = 11.84HH6 pKa = 5.89HH7 pKa = 6.51HH8 pKa = 5.09TTTTTTTRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84GLFSSRR24 pKa = 11.84RR25 pKa = 11.84QPVVVHH31 pKa = 5.1HH32 pKa = 6.06QKK34 pKa = 10.13RR35 pKa = 11.84HH36 pKa = 4.31ATLGDD41 pKa = 4.03KK42 pKa = 10.67ISGAFLKK49 pKa = 10.97LKK51 pKa = 10.56GSLTGRR57 pKa = 11.84PGEE60 pKa = 4.13KK61 pKa = 9.9AAGTRR66 pKa = 11.84RR67 pKa = 11.84MRR69 pKa = 11.84GTDD72 pKa = 2.74GRR74 pKa = 11.84GSHH77 pKa = 5.71RR78 pKa = 11.84RR79 pKa = 11.84RR80 pKa = 11.84RR81 pKa = 11.84FFF83 pKa = 4.33
Molecular weight: 9.47 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3911874 |
66 |
10679 |
442.9 |
48.92 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.1 ± 0.026 | 1.07 ± 0.008 |
6.047 ± 0.02 | 6.422 ± 0.03 |
3.849 ± 0.017 | 7.343 ± 0.025 |
2.138 ± 0.011 | 5.117 ± 0.017 |
5.337 ± 0.025 | 8.707 ± 0.027 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.263 ± 0.011 | 3.676 ± 0.014 |
5.489 ± 0.024 | 3.793 ± 0.017 |
5.581 ± 0.02 | 7.316 ± 0.023 |
5.857 ± 0.02 | 6.508 ± 0.017 |
1.491 ± 0.011 | 2.896 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
