
Pelargonium ringspot virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Tolucaviricetes; Tolivirales; Tombusviridae; Procedovirinae; Pelarspovirus
Average proteome isoelectric point is 8.55
Get precalculated fractions of proteins
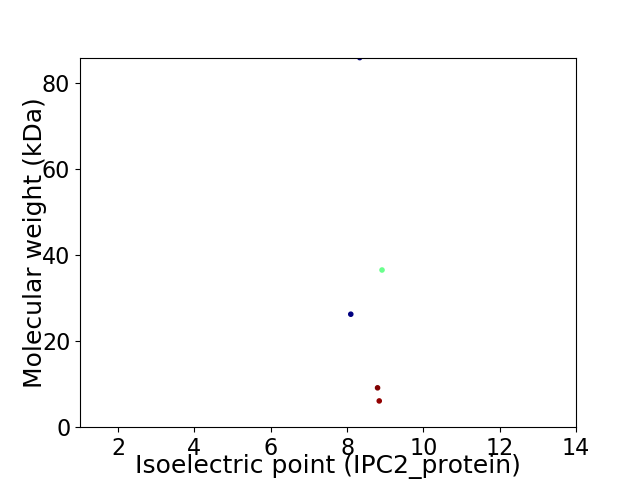
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q911J5|Q911J5_9TOMB Capsid protein OS=Pelargonium ringspot virus OX=167020 GN=p37 PE=3 SV=1
MM1 pKa = 7.4TDD3 pKa = 3.07VVKK6 pKa = 10.78FGVWAGFYY14 pKa = 9.96LAKK17 pKa = 10.25EE18 pKa = 3.99LALLGPEE25 pKa = 4.33LASITLSSLTPNYY38 pKa = 10.49NGLLTSIGLPTVSSIPTASPPIDD61 pKa = 3.23LVLVNDD67 pKa = 4.83GVLNTPNLEE76 pKa = 4.24LEE78 pKa = 4.61GEE80 pKa = 4.25LVPLLEE86 pKa = 5.17EE87 pKa = 4.85VITDD91 pKa = 4.26PKK93 pKa = 10.43TGTITKK99 pKa = 9.61AVKK102 pKa = 8.99TRR104 pKa = 11.84RR105 pKa = 11.84PKK107 pKa = 10.73SKK109 pKa = 7.87TTFSCILAAEE119 pKa = 4.56TKK121 pKa = 10.1NHH123 pKa = 6.92FGGLPRR129 pKa = 11.84ATRR132 pKa = 11.84ANEE135 pKa = 3.79LSVMKK140 pKa = 10.53HH141 pKa = 4.93LVNRR145 pKa = 11.84CKK147 pKa = 10.4EE148 pKa = 4.28CKK150 pKa = 9.08LTALQTRR157 pKa = 11.84EE158 pKa = 3.76VSAKK162 pKa = 10.3AFSLVFTPDD171 pKa = 2.78SHH173 pKa = 8.07DD174 pKa = 3.34KK175 pKa = 10.89FIYY178 pKa = 10.27EE179 pKa = 4.59FLNSDD184 pKa = 3.01ITFEE188 pKa = 4.53RR189 pKa = 11.84RR190 pKa = 11.84CDD192 pKa = 3.57YY193 pKa = 11.04LKK195 pKa = 10.56SQRR198 pKa = 11.84VDD200 pKa = 3.55SCWLRR205 pKa = 11.84LLQNPFGKK213 pKa = 10.16KK214 pKa = 6.4RR215 pKa = 11.84WKK217 pKa = 10.72AVVCRR222 pKa = 11.84LMGMGVQEE230 pKa = 4.21AYY232 pKa = 10.38EE233 pKa = 4.07FVKK236 pKa = 10.97
MM1 pKa = 7.4TDD3 pKa = 3.07VVKK6 pKa = 10.78FGVWAGFYY14 pKa = 9.96LAKK17 pKa = 10.25EE18 pKa = 3.99LALLGPEE25 pKa = 4.33LASITLSSLTPNYY38 pKa = 10.49NGLLTSIGLPTVSSIPTASPPIDD61 pKa = 3.23LVLVNDD67 pKa = 4.83GVLNTPNLEE76 pKa = 4.24LEE78 pKa = 4.61GEE80 pKa = 4.25LVPLLEE86 pKa = 5.17EE87 pKa = 4.85VITDD91 pKa = 4.26PKK93 pKa = 10.43TGTITKK99 pKa = 9.61AVKK102 pKa = 8.99TRR104 pKa = 11.84RR105 pKa = 11.84PKK107 pKa = 10.73SKK109 pKa = 7.87TTFSCILAAEE119 pKa = 4.56TKK121 pKa = 10.1NHH123 pKa = 6.92FGGLPRR129 pKa = 11.84ATRR132 pKa = 11.84ANEE135 pKa = 3.79LSVMKK140 pKa = 10.53HH141 pKa = 4.93LVNRR145 pKa = 11.84CKK147 pKa = 10.4EE148 pKa = 4.28CKK150 pKa = 9.08LTALQTRR157 pKa = 11.84EE158 pKa = 3.76VSAKK162 pKa = 10.3AFSLVFTPDD171 pKa = 2.78SHH173 pKa = 8.07DD174 pKa = 3.34KK175 pKa = 10.89FIYY178 pKa = 10.27EE179 pKa = 4.59FLNSDD184 pKa = 3.01ITFEE188 pKa = 4.53RR189 pKa = 11.84RR190 pKa = 11.84CDD192 pKa = 3.57YY193 pKa = 11.04LKK195 pKa = 10.56SQRR198 pKa = 11.84VDD200 pKa = 3.55SCWLRR205 pKa = 11.84LLQNPFGKK213 pKa = 10.16KK214 pKa = 6.4RR215 pKa = 11.84WKK217 pKa = 10.72AVVCRR222 pKa = 11.84LMGMGVQEE230 pKa = 4.21AYY232 pKa = 10.38EE233 pKa = 4.07FVKK236 pKa = 10.97
Molecular weight: 26.27 kDa
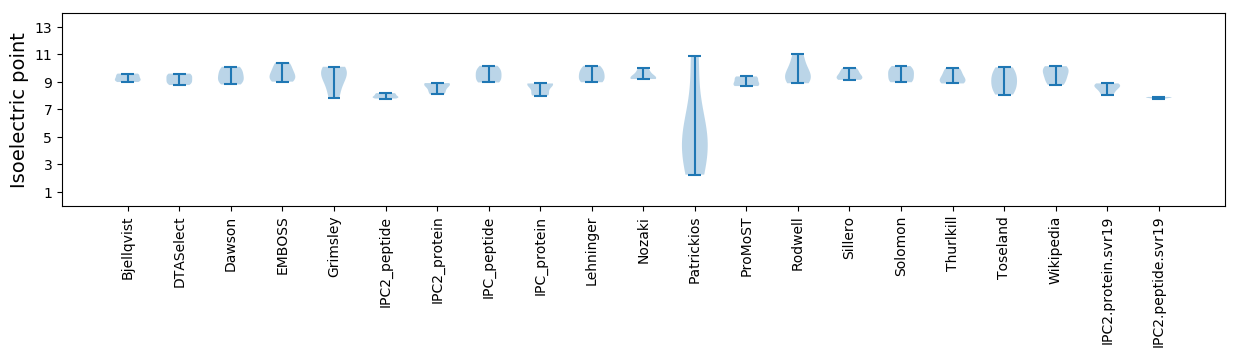
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q911J5|Q911J5_9TOMB Capsid protein OS=Pelargonium ringspot virus OX=167020 GN=p37 PE=3 SV=1
MM1 pKa = 7.74ASNSPIVLAAANRR14 pKa = 11.84GEE16 pKa = 4.14VWAVKK21 pKa = 10.24LKK23 pKa = 9.99QSGWKK28 pKa = 8.19TLSKK32 pKa = 10.15AQKK35 pKa = 9.73AAARR39 pKa = 11.84AQGIGAAPLSQTVPKK54 pKa = 7.67TTRR57 pKa = 11.84IIQGNPTFSRR67 pKa = 11.84RR68 pKa = 11.84SRR70 pKa = 11.84DD71 pKa = 3.43APGNAQKK78 pKa = 11.43SMTIVKK84 pKa = 10.09SEE86 pKa = 3.84FLGEE90 pKa = 4.02VSADD94 pKa = 3.9GAIHH98 pKa = 7.34TYY100 pKa = 9.78TLDD103 pKa = 3.45PRR105 pKa = 11.84NVRR108 pKa = 11.84TFPHH112 pKa = 6.46ISDD115 pKa = 4.47LARR118 pKa = 11.84GFNKK122 pKa = 10.4YY123 pKa = 10.36KK124 pKa = 10.77FGDD127 pKa = 3.53LKK129 pKa = 10.95VRR131 pKa = 11.84YY132 pKa = 8.35SAKK135 pKa = 10.08VQEE138 pKa = 4.74SGCGVCIAFTPDD150 pKa = 3.13SSDD153 pKa = 3.5PKK155 pKa = 10.59PKK157 pKa = 10.71GKK159 pKa = 10.15FDD161 pKa = 4.29LYY163 pKa = 11.22SLGSRR168 pKa = 11.84AEE170 pKa = 4.28GAAHH174 pKa = 7.36RR175 pKa = 11.84NLLYY179 pKa = 9.88TVPIDD184 pKa = 3.19KK185 pKa = 10.43SQRR188 pKa = 11.84FLRR191 pKa = 11.84DD192 pKa = 3.05CATEE196 pKa = 3.8NSKK199 pKa = 11.2DD200 pKa = 3.35VDD202 pKa = 3.72AGSLFVLVDD211 pKa = 3.72GQHH214 pKa = 6.8DD215 pKa = 3.99GRR217 pKa = 11.84VGEE220 pKa = 4.86LFLEE224 pKa = 4.5VTIVLSQPTYY234 pKa = 9.22NQRR237 pKa = 11.84ATQLLAGTTHH247 pKa = 7.3RR248 pKa = 11.84DD249 pKa = 2.95GPTFVKK255 pKa = 10.33AVKK258 pKa = 9.91SANTVSLTFQAAGNYY273 pKa = 8.89LVSTYY278 pKa = 11.09SSLLEE283 pKa = 3.98RR284 pKa = 11.84VGRR287 pKa = 11.84LALGEE292 pKa = 4.34AEE294 pKa = 4.09QSQVDD299 pKa = 4.06SSTHH303 pKa = 5.24SSNVVEE309 pKa = 4.83ALVAAPGGSIVYY321 pKa = 8.83YY322 pKa = 10.45FKK324 pKa = 10.83TPDD327 pKa = 3.2QMSFRR332 pKa = 11.84AYY334 pKa = 8.46VCRR337 pKa = 11.84MM338 pKa = 3.31
MM1 pKa = 7.74ASNSPIVLAAANRR14 pKa = 11.84GEE16 pKa = 4.14VWAVKK21 pKa = 10.24LKK23 pKa = 9.99QSGWKK28 pKa = 8.19TLSKK32 pKa = 10.15AQKK35 pKa = 9.73AAARR39 pKa = 11.84AQGIGAAPLSQTVPKK54 pKa = 7.67TTRR57 pKa = 11.84IIQGNPTFSRR67 pKa = 11.84RR68 pKa = 11.84SRR70 pKa = 11.84DD71 pKa = 3.43APGNAQKK78 pKa = 11.43SMTIVKK84 pKa = 10.09SEE86 pKa = 3.84FLGEE90 pKa = 4.02VSADD94 pKa = 3.9GAIHH98 pKa = 7.34TYY100 pKa = 9.78TLDD103 pKa = 3.45PRR105 pKa = 11.84NVRR108 pKa = 11.84TFPHH112 pKa = 6.46ISDD115 pKa = 4.47LARR118 pKa = 11.84GFNKK122 pKa = 10.4YY123 pKa = 10.36KK124 pKa = 10.77FGDD127 pKa = 3.53LKK129 pKa = 10.95VRR131 pKa = 11.84YY132 pKa = 8.35SAKK135 pKa = 10.08VQEE138 pKa = 4.74SGCGVCIAFTPDD150 pKa = 3.13SSDD153 pKa = 3.5PKK155 pKa = 10.59PKK157 pKa = 10.71GKK159 pKa = 10.15FDD161 pKa = 4.29LYY163 pKa = 11.22SLGSRR168 pKa = 11.84AEE170 pKa = 4.28GAAHH174 pKa = 7.36RR175 pKa = 11.84NLLYY179 pKa = 9.88TVPIDD184 pKa = 3.19KK185 pKa = 10.43SQRR188 pKa = 11.84FLRR191 pKa = 11.84DD192 pKa = 3.05CATEE196 pKa = 3.8NSKK199 pKa = 11.2DD200 pKa = 3.35VDD202 pKa = 3.72AGSLFVLVDD211 pKa = 3.72GQHH214 pKa = 6.8DD215 pKa = 3.99GRR217 pKa = 11.84VGEE220 pKa = 4.86LFLEE224 pKa = 4.5VTIVLSQPTYY234 pKa = 9.22NQRR237 pKa = 11.84ATQLLAGTTHH247 pKa = 7.3RR248 pKa = 11.84DD249 pKa = 2.95GPTFVKK255 pKa = 10.33AVKK258 pKa = 9.91SANTVSLTFQAAGNYY273 pKa = 8.89LVSTYY278 pKa = 11.09SSLLEE283 pKa = 3.98RR284 pKa = 11.84VGRR287 pKa = 11.84LALGEE292 pKa = 4.34AEE294 pKa = 4.09QSQVDD299 pKa = 4.06SSTHH303 pKa = 5.24SSNVVEE309 pKa = 4.83ALVAAPGGSIVYY321 pKa = 8.83YY322 pKa = 10.45FKK324 pKa = 10.83TPDD327 pKa = 3.2QMSFRR332 pKa = 11.84AYY334 pKa = 8.46VCRR337 pKa = 11.84MM338 pKa = 3.31
Molecular weight: 36.55 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1477 |
58 |
762 |
295.4 |
32.8 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.703 ± 1.292 | 1.828 ± 0.315 |
4.333 ± 0.316 | 5.213 ± 0.909 |
4.333 ± 0.269 | 6.838 ± 0.421 |
2.099 ± 0.481 | 4.265 ± 0.448 |
6.838 ± 0.415 | 10.088 ± 1.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.828 ± 0.291 | 3.995 ± 0.181 |
5.146 ± 0.273 | 2.911 ± 0.592 |
5.619 ± 0.734 | 8.125 ± 1.485 |
6.5 ± 0.54 | 8.734 ± 0.43 |
1.219 ± 0.232 | 3.318 ± 0.358 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
