
Nkolbisson virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alpharhabdovirinae; Ledantevirus; Nkolbisson ledantevirus
Average proteome isoelectric point is 6.0
Get precalculated fractions of proteins
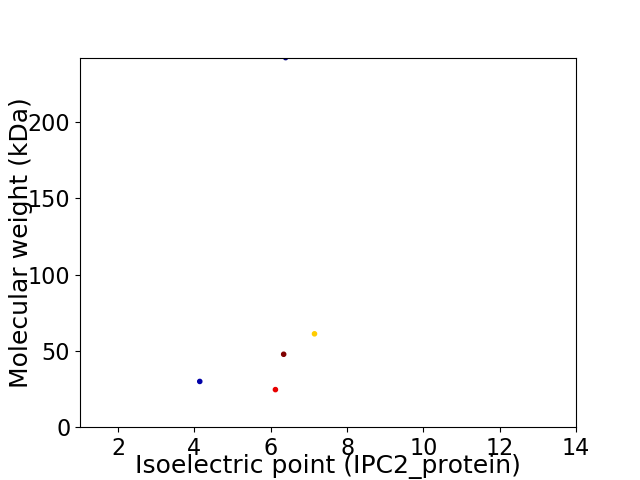
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0D3R1P3|A0A0D3R1P3_9RHAB Glycoprotein OS=Nkolbisson virus OX=380442 PE=4 SV=1
MM1 pKa = 7.23SRR3 pKa = 11.84QRR5 pKa = 11.84LRR7 pKa = 11.84EE8 pKa = 3.33IAGRR12 pKa = 11.84YY13 pKa = 9.19DD14 pKa = 3.46YY15 pKa = 11.8SRR17 pKa = 11.84VAKK20 pKa = 10.21NLEE23 pKa = 3.79GLEE26 pKa = 3.83EE27 pKa = 4.45DD28 pKa = 3.93EE29 pKa = 5.1NEE31 pKa = 4.43KK32 pKa = 10.66EE33 pKa = 4.04LEE35 pKa = 4.19AAAGTIPANIVEE47 pKa = 4.73EE48 pKa = 4.39PDD50 pKa = 3.43SSEE53 pKa = 5.48DD54 pKa = 3.61EE55 pKa = 4.79ASDD58 pKa = 3.64GDD60 pKa = 3.9EE61 pKa = 4.51EE62 pKa = 4.05EE63 pKa = 4.47LEE65 pKa = 4.25RR66 pKa = 11.84IFRR69 pKa = 11.84EE70 pKa = 4.18DD71 pKa = 3.2EE72 pKa = 3.94KK73 pKa = 11.6SEE75 pKa = 4.51DD76 pKa = 3.7EE77 pKa = 4.69LEE79 pKa = 4.22NDD81 pKa = 3.31HH82 pKa = 7.39FEE84 pKa = 4.45LSLEE88 pKa = 4.17AEE90 pKa = 4.43EE91 pKa = 4.28EE92 pKa = 4.39AEE94 pKa = 4.92LEE96 pKa = 4.23SSSDD100 pKa = 3.23KK101 pKa = 11.07TIILPPMPPLSGLRR115 pKa = 11.84YY116 pKa = 9.33QDD118 pKa = 3.34EE119 pKa = 4.52LEE121 pKa = 4.44NIILHH126 pKa = 5.32YY127 pKa = 10.78VSTSLASVGWYY138 pKa = 10.28LNPQQIEE145 pKa = 4.09KK146 pKa = 10.95GEE148 pKa = 4.13GGLRR152 pKa = 11.84VGINKK157 pKa = 8.32ITDD160 pKa = 3.57NQPLSSLDD168 pKa = 3.2TTAPEE173 pKa = 3.78EE174 pKa = 4.22SLDD177 pKa = 4.01VVIPDD182 pKa = 4.1DD183 pKa = 4.82LIKK186 pKa = 10.94EE187 pKa = 4.29EE188 pKa = 4.58EE189 pKa = 4.36DD190 pKa = 3.58TVTRR194 pKa = 11.84PDD196 pKa = 2.96VDD198 pKa = 3.34KK199 pKa = 11.23LFRR202 pKa = 11.84EE203 pKa = 4.7GLSFTRR209 pKa = 11.84KK210 pKa = 8.4GNKK213 pKa = 9.81GIIEE217 pKa = 4.45VNSEE221 pKa = 3.94TPGWSKK227 pKa = 10.98EE228 pKa = 3.85LLEE231 pKa = 4.93FATTMFGNRR240 pKa = 11.84QDD242 pKa = 3.77QIDD245 pKa = 4.43YY246 pKa = 10.9ILDD249 pKa = 3.77EE250 pKa = 5.1LEE252 pKa = 4.58LLPMIKK258 pKa = 9.87RR259 pKa = 11.84LCIYY263 pKa = 9.7PP264 pKa = 3.72
MM1 pKa = 7.23SRR3 pKa = 11.84QRR5 pKa = 11.84LRR7 pKa = 11.84EE8 pKa = 3.33IAGRR12 pKa = 11.84YY13 pKa = 9.19DD14 pKa = 3.46YY15 pKa = 11.8SRR17 pKa = 11.84VAKK20 pKa = 10.21NLEE23 pKa = 3.79GLEE26 pKa = 3.83EE27 pKa = 4.45DD28 pKa = 3.93EE29 pKa = 5.1NEE31 pKa = 4.43KK32 pKa = 10.66EE33 pKa = 4.04LEE35 pKa = 4.19AAAGTIPANIVEE47 pKa = 4.73EE48 pKa = 4.39PDD50 pKa = 3.43SSEE53 pKa = 5.48DD54 pKa = 3.61EE55 pKa = 4.79ASDD58 pKa = 3.64GDD60 pKa = 3.9EE61 pKa = 4.51EE62 pKa = 4.05EE63 pKa = 4.47LEE65 pKa = 4.25RR66 pKa = 11.84IFRR69 pKa = 11.84EE70 pKa = 4.18DD71 pKa = 3.2EE72 pKa = 3.94KK73 pKa = 11.6SEE75 pKa = 4.51DD76 pKa = 3.7EE77 pKa = 4.69LEE79 pKa = 4.22NDD81 pKa = 3.31HH82 pKa = 7.39FEE84 pKa = 4.45LSLEE88 pKa = 4.17AEE90 pKa = 4.43EE91 pKa = 4.28EE92 pKa = 4.39AEE94 pKa = 4.92LEE96 pKa = 4.23SSSDD100 pKa = 3.23KK101 pKa = 11.07TIILPPMPPLSGLRR115 pKa = 11.84YY116 pKa = 9.33QDD118 pKa = 3.34EE119 pKa = 4.52LEE121 pKa = 4.44NIILHH126 pKa = 5.32YY127 pKa = 10.78VSTSLASVGWYY138 pKa = 10.28LNPQQIEE145 pKa = 4.09KK146 pKa = 10.95GEE148 pKa = 4.13GGLRR152 pKa = 11.84VGINKK157 pKa = 8.32ITDD160 pKa = 3.57NQPLSSLDD168 pKa = 3.2TTAPEE173 pKa = 3.78EE174 pKa = 4.22SLDD177 pKa = 4.01VVIPDD182 pKa = 4.1DD183 pKa = 4.82LIKK186 pKa = 10.94EE187 pKa = 4.29EE188 pKa = 4.58EE189 pKa = 4.36DD190 pKa = 3.58TVTRR194 pKa = 11.84PDD196 pKa = 2.96VDD198 pKa = 3.34KK199 pKa = 11.23LFRR202 pKa = 11.84EE203 pKa = 4.7GLSFTRR209 pKa = 11.84KK210 pKa = 8.4GNKK213 pKa = 9.81GIIEE217 pKa = 4.45VNSEE221 pKa = 3.94TPGWSKK227 pKa = 10.98EE228 pKa = 3.85LLEE231 pKa = 4.93FATTMFGNRR240 pKa = 11.84QDD242 pKa = 3.77QIDD245 pKa = 4.43YY246 pKa = 10.9ILDD249 pKa = 3.77EE250 pKa = 5.1LEE252 pKa = 4.58LLPMIKK258 pKa = 9.87RR259 pKa = 11.84LCIYY263 pKa = 9.7PP264 pKa = 3.72
Molecular weight: 29.97 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0D3R205|A0A0D3R205_9RHAB Nucleocapsid protein OS=Nkolbisson virus OX=380442 PE=4 SV=1
MM1 pKa = 7.06QLSLLMVMASLAQSVTGRR19 pKa = 11.84AAGRR23 pKa = 11.84EE24 pKa = 3.97LHH26 pKa = 6.97DD27 pKa = 4.28VIGFVPDD34 pKa = 3.47TDD36 pKa = 3.75TLQWKK41 pKa = 8.1PANLDD46 pKa = 3.38TLRR49 pKa = 11.84CPEE52 pKa = 4.4AADD55 pKa = 4.45PGPDD59 pKa = 3.38DD60 pKa = 3.96GAVVEE65 pKa = 4.46KK66 pKa = 10.2WVISRR71 pKa = 11.84PRR73 pKa = 11.84VNTFLGIKK81 pKa = 10.25GYY83 pKa = 10.14LCHH86 pKa = 5.71YY87 pKa = 8.43AKK89 pKa = 10.19WVTRR93 pKa = 11.84CEE95 pKa = 4.19YY96 pKa = 9.51TWYY99 pKa = 10.45LSKK102 pKa = 10.02TISRR106 pKa = 11.84SIQPLDD112 pKa = 3.53VTEE115 pKa = 5.09SGCQDD120 pKa = 2.81AFKK123 pKa = 10.47EE124 pKa = 4.04YY125 pKa = 10.61DD126 pKa = 3.15AGRR129 pKa = 11.84LVPGSFPPEE138 pKa = 3.44ACYY141 pKa = 9.05WASTNDD147 pKa = 3.9EE148 pKa = 4.26TVHH151 pKa = 6.83AYY153 pKa = 8.77TITPHH158 pKa = 5.6EE159 pKa = 4.74VIYY162 pKa = 10.71DD163 pKa = 3.99PYY165 pKa = 11.57SNAKK169 pKa = 9.31LDD171 pKa = 4.09HH172 pKa = 6.56IFLHH176 pKa = 6.61GSCNKK181 pKa = 9.9DD182 pKa = 3.17FCEE185 pKa = 4.24TSHH188 pKa = 7.58DD189 pKa = 3.75STMWIAKK196 pKa = 9.66RR197 pKa = 11.84KK198 pKa = 10.3GEE200 pKa = 3.99GSVCQFVGRR209 pKa = 11.84EE210 pKa = 3.85DD211 pKa = 3.54VEE213 pKa = 4.39IIEE216 pKa = 4.34GAKK219 pKa = 10.04FSLRR223 pKa = 11.84KK224 pKa = 9.27FQTKK228 pKa = 7.76VWLRR232 pKa = 11.84GPSLHH237 pKa = 6.99NIPLDD242 pKa = 3.69KK243 pKa = 10.67LCKK246 pKa = 9.22IDD248 pKa = 3.94FCGKK252 pKa = 8.17TGYY255 pKa = 10.36RR256 pKa = 11.84NQQGIWFSIDD266 pKa = 2.78RR267 pKa = 11.84VSWSKK272 pKa = 10.74NHH274 pKa = 6.38SLPIRR279 pKa = 11.84QKK281 pKa = 11.24AVGCKK286 pKa = 9.37PGEE289 pKa = 4.09NVAVVDD295 pKa = 4.14TNLPDD300 pKa = 3.92SDD302 pKa = 4.64LEE304 pKa = 4.17ATIEE308 pKa = 3.98EE309 pKa = 4.71MMWDD313 pKa = 3.99MNCLNALEE321 pKa = 4.96TIQHH325 pKa = 5.67HH326 pKa = 6.42KK327 pKa = 10.35KK328 pKa = 10.75VSLHH332 pKa = 6.82DD333 pKa = 4.72LYY335 pKa = 11.26QIAPQKK341 pKa = 10.4SGPGLAYY348 pKa = 10.61RR349 pKa = 11.84LNDD352 pKa = 3.18GHH354 pKa = 8.81LEE356 pKa = 4.17VAQAHH361 pKa = 5.54YY362 pKa = 10.3WSVQHH367 pKa = 6.36PKK369 pKa = 10.1RR370 pKa = 11.84GKK372 pKa = 10.35LSRR375 pKa = 11.84TCLGARR381 pKa = 11.84QADD384 pKa = 3.98EE385 pKa = 4.08TSSCVKK391 pKa = 8.48WNKK394 pKa = 6.02WTHH397 pKa = 4.47VANNTYY403 pKa = 10.19HH404 pKa = 6.67AVNGITEE411 pKa = 4.08VDD413 pKa = 3.76GQILFPKK420 pKa = 10.03RR421 pKa = 11.84RR422 pKa = 11.84VLHH425 pKa = 6.24RR426 pKa = 11.84KK427 pKa = 9.2YY428 pKa = 10.61DD429 pKa = 3.52PEE431 pKa = 4.04YY432 pKa = 10.31ATRR435 pKa = 11.84QSLRR439 pKa = 11.84FIKK442 pKa = 10.58HH443 pKa = 4.53PVIDD447 pKa = 3.89KK448 pKa = 11.15FEE450 pKa = 4.8DD451 pKa = 3.41KK452 pKa = 10.17ATEE455 pKa = 4.34NIEE458 pKa = 4.11HH459 pKa = 7.29KK460 pKa = 10.73DD461 pKa = 3.57IISKK465 pKa = 10.34DD466 pKa = 3.58VNAGDD471 pKa = 5.2LVGNWVVVAEE481 pKa = 4.83GKK483 pKa = 9.6LSEE486 pKa = 4.2WFGGLGKK493 pKa = 10.7SIISVVSLIVGLLVLYY509 pKa = 9.43VIVKK513 pKa = 8.87LCLYY517 pKa = 8.55FRR519 pKa = 11.84PKK521 pKa = 10.6AKK523 pKa = 10.34EE524 pKa = 3.88KK525 pKa = 10.63IGDD528 pKa = 3.63MEE530 pKa = 4.35MKK532 pKa = 10.23LKK534 pKa = 10.46KK535 pKa = 9.98RR536 pKa = 11.84SEE538 pKa = 4.33SFGG541 pKa = 3.45
MM1 pKa = 7.06QLSLLMVMASLAQSVTGRR19 pKa = 11.84AAGRR23 pKa = 11.84EE24 pKa = 3.97LHH26 pKa = 6.97DD27 pKa = 4.28VIGFVPDD34 pKa = 3.47TDD36 pKa = 3.75TLQWKK41 pKa = 8.1PANLDD46 pKa = 3.38TLRR49 pKa = 11.84CPEE52 pKa = 4.4AADD55 pKa = 4.45PGPDD59 pKa = 3.38DD60 pKa = 3.96GAVVEE65 pKa = 4.46KK66 pKa = 10.2WVISRR71 pKa = 11.84PRR73 pKa = 11.84VNTFLGIKK81 pKa = 10.25GYY83 pKa = 10.14LCHH86 pKa = 5.71YY87 pKa = 8.43AKK89 pKa = 10.19WVTRR93 pKa = 11.84CEE95 pKa = 4.19YY96 pKa = 9.51TWYY99 pKa = 10.45LSKK102 pKa = 10.02TISRR106 pKa = 11.84SIQPLDD112 pKa = 3.53VTEE115 pKa = 5.09SGCQDD120 pKa = 2.81AFKK123 pKa = 10.47EE124 pKa = 4.04YY125 pKa = 10.61DD126 pKa = 3.15AGRR129 pKa = 11.84LVPGSFPPEE138 pKa = 3.44ACYY141 pKa = 9.05WASTNDD147 pKa = 3.9EE148 pKa = 4.26TVHH151 pKa = 6.83AYY153 pKa = 8.77TITPHH158 pKa = 5.6EE159 pKa = 4.74VIYY162 pKa = 10.71DD163 pKa = 3.99PYY165 pKa = 11.57SNAKK169 pKa = 9.31LDD171 pKa = 4.09HH172 pKa = 6.56IFLHH176 pKa = 6.61GSCNKK181 pKa = 9.9DD182 pKa = 3.17FCEE185 pKa = 4.24TSHH188 pKa = 7.58DD189 pKa = 3.75STMWIAKK196 pKa = 9.66RR197 pKa = 11.84KK198 pKa = 10.3GEE200 pKa = 3.99GSVCQFVGRR209 pKa = 11.84EE210 pKa = 3.85DD211 pKa = 3.54VEE213 pKa = 4.39IIEE216 pKa = 4.34GAKK219 pKa = 10.04FSLRR223 pKa = 11.84KK224 pKa = 9.27FQTKK228 pKa = 7.76VWLRR232 pKa = 11.84GPSLHH237 pKa = 6.99NIPLDD242 pKa = 3.69KK243 pKa = 10.67LCKK246 pKa = 9.22IDD248 pKa = 3.94FCGKK252 pKa = 8.17TGYY255 pKa = 10.36RR256 pKa = 11.84NQQGIWFSIDD266 pKa = 2.78RR267 pKa = 11.84VSWSKK272 pKa = 10.74NHH274 pKa = 6.38SLPIRR279 pKa = 11.84QKK281 pKa = 11.24AVGCKK286 pKa = 9.37PGEE289 pKa = 4.09NVAVVDD295 pKa = 4.14TNLPDD300 pKa = 3.92SDD302 pKa = 4.64LEE304 pKa = 4.17ATIEE308 pKa = 3.98EE309 pKa = 4.71MMWDD313 pKa = 3.99MNCLNALEE321 pKa = 4.96TIQHH325 pKa = 5.67HH326 pKa = 6.42KK327 pKa = 10.35KK328 pKa = 10.75VSLHH332 pKa = 6.82DD333 pKa = 4.72LYY335 pKa = 11.26QIAPQKK341 pKa = 10.4SGPGLAYY348 pKa = 10.61RR349 pKa = 11.84LNDD352 pKa = 3.18GHH354 pKa = 8.81LEE356 pKa = 4.17VAQAHH361 pKa = 5.54YY362 pKa = 10.3WSVQHH367 pKa = 6.36PKK369 pKa = 10.1RR370 pKa = 11.84GKK372 pKa = 10.35LSRR375 pKa = 11.84TCLGARR381 pKa = 11.84QADD384 pKa = 3.98EE385 pKa = 4.08TSSCVKK391 pKa = 8.48WNKK394 pKa = 6.02WTHH397 pKa = 4.47VANNTYY403 pKa = 10.19HH404 pKa = 6.67AVNGITEE411 pKa = 4.08VDD413 pKa = 3.76GQILFPKK420 pKa = 10.03RR421 pKa = 11.84RR422 pKa = 11.84VLHH425 pKa = 6.24RR426 pKa = 11.84KK427 pKa = 9.2YY428 pKa = 10.61DD429 pKa = 3.52PEE431 pKa = 4.04YY432 pKa = 10.31ATRR435 pKa = 11.84QSLRR439 pKa = 11.84FIKK442 pKa = 10.58HH443 pKa = 4.53PVIDD447 pKa = 3.89KK448 pKa = 11.15FEE450 pKa = 4.8DD451 pKa = 3.41KK452 pKa = 10.17ATEE455 pKa = 4.34NIEE458 pKa = 4.11HH459 pKa = 7.29KK460 pKa = 10.73DD461 pKa = 3.57IISKK465 pKa = 10.34DD466 pKa = 3.58VNAGDD471 pKa = 5.2LVGNWVVVAEE481 pKa = 4.83GKK483 pKa = 9.6LSEE486 pKa = 4.2WFGGLGKK493 pKa = 10.7SIISVVSLIVGLLVLYY509 pKa = 9.43VIVKK513 pKa = 8.87LCLYY517 pKa = 8.55FRR519 pKa = 11.84PKK521 pKa = 10.6AKK523 pKa = 10.34EE524 pKa = 3.88KK525 pKa = 10.63IGDD528 pKa = 3.63MEE530 pKa = 4.35MKK532 pKa = 10.23LKK534 pKa = 10.46KK535 pKa = 9.98RR536 pKa = 11.84SEE538 pKa = 4.33SFGG541 pKa = 3.45
Molecular weight: 61.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3550 |
210 |
2114 |
710.0 |
81.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.732 ± 0.849 | 2.113 ± 0.254 |
6.254 ± 0.277 | 7.183 ± 1.226 |
3.915 ± 0.417 | 6.0 ± 0.441 |
2.704 ± 0.315 | 6.394 ± 0.45 |
6.394 ± 0.346 | 9.972 ± 0.395 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.423 ± 0.294 | 4.197 ± 0.238 |
4.563 ± 0.537 | 3.127 ± 0.083 |
5.746 ± 0.331 | 7.211 ± 0.764 |
5.324 ± 0.186 | 6.31 ± 0.532 |
1.859 ± 0.225 | 3.577 ± 0.242 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
