
Torque teno sus virus 1 (isolate Sd-TTV31)
Taxonomy: Viruses; Anelloviridae; Iotatorquevirus; Torque teno sus virus 1a
Average proteome isoelectric point is 6.27
Get precalculated fractions of proteins
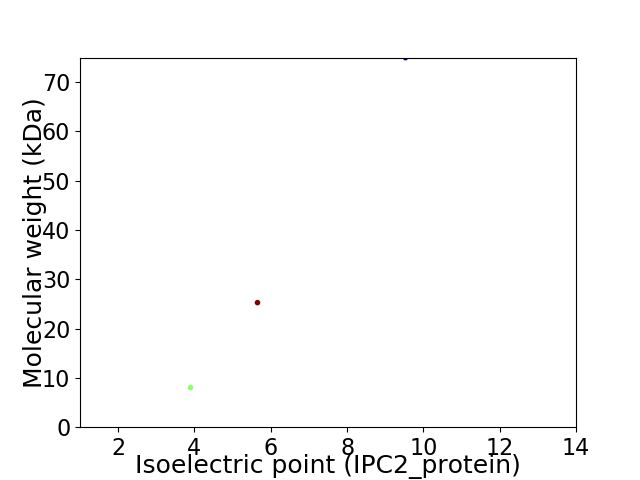
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q8QVM1|ORF3_TTVI1 Uncharacterized ORF3 protein OS=Torque teno sus virus 1 (isolate Sd-TTV31) OX=766190 GN=ORF1 PE=4 SV=1
MM1 pKa = 7.54KK2 pKa = 10.41EE3 pKa = 3.35KK4 pKa = 10.56DD5 pKa = 3.44YY6 pKa = 11.27WEE8 pKa = 4.4EE9 pKa = 3.78AWLTSCTSIHH19 pKa = 7.02DD20 pKa = 3.74HH21 pKa = 6.84HH22 pKa = 7.45CDD24 pKa = 3.28CGSWRR29 pKa = 11.84DD30 pKa = 4.0HH31 pKa = 6.31LWTLCALDD39 pKa = 4.96DD40 pKa = 4.87ADD42 pKa = 4.9LAAAADD48 pKa = 3.67IIEE51 pKa = 4.5RR52 pKa = 11.84EE53 pKa = 4.08EE54 pKa = 4.81ADD56 pKa = 3.34GGEE59 pKa = 4.28DD60 pKa = 3.6FGFVDD65 pKa = 5.31GDD67 pKa = 3.79PGDD70 pKa = 3.85AGGG73 pKa = 4.2
MM1 pKa = 7.54KK2 pKa = 10.41EE3 pKa = 3.35KK4 pKa = 10.56DD5 pKa = 3.44YY6 pKa = 11.27WEE8 pKa = 4.4EE9 pKa = 3.78AWLTSCTSIHH19 pKa = 7.02DD20 pKa = 3.74HH21 pKa = 6.84HH22 pKa = 7.45CDD24 pKa = 3.28CGSWRR29 pKa = 11.84DD30 pKa = 4.0HH31 pKa = 6.31LWTLCALDD39 pKa = 4.96DD40 pKa = 4.87ADD42 pKa = 4.9LAAAADD48 pKa = 3.67IIEE51 pKa = 4.5RR52 pKa = 11.84EE53 pKa = 4.08EE54 pKa = 4.81ADD56 pKa = 3.34GGEE59 pKa = 4.28DD60 pKa = 3.6FGFVDD65 pKa = 5.31GDD67 pKa = 3.79PGDD70 pKa = 3.85AGGG73 pKa = 4.2
Molecular weight: 8.04 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q8QVM0|ORF2_TTVI1 Uncharacterized ORF2 protein OS=Torque teno sus virus 1 (isolate Sd-TTV31) OX=766190 GN=ORF1 PE=4 SV=1
MM1 pKa = 7.79RR2 pKa = 11.84FRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84FGRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84YY14 pKa = 7.87YY15 pKa = 9.54RR16 pKa = 11.84KK17 pKa = 9.61RR18 pKa = 11.84RR19 pKa = 11.84GGWRR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84FRR27 pKa = 11.84IRR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84PWRR35 pKa = 11.84RR36 pKa = 11.84WRR38 pKa = 11.84VRR40 pKa = 11.84RR41 pKa = 11.84WRR43 pKa = 11.84RR44 pKa = 11.84SVFRR48 pKa = 11.84RR49 pKa = 11.84RR50 pKa = 11.84GRR52 pKa = 11.84RR53 pKa = 11.84ARR55 pKa = 11.84PYY57 pKa = 10.21RR58 pKa = 11.84ISAWNPKK65 pKa = 7.44VLRR68 pKa = 11.84NCRR71 pKa = 11.84ITGWWPVIQCMDD83 pKa = 2.9GMEE86 pKa = 4.32WIKK89 pKa = 10.41YY90 pKa = 9.99KK91 pKa = 10.93PMDD94 pKa = 3.88LRR96 pKa = 11.84VEE98 pKa = 4.09ANRR101 pKa = 11.84IFDD104 pKa = 3.72KK105 pKa = 10.88QGSKK109 pKa = 10.07IEE111 pKa = 4.16TEE113 pKa = 3.91QMGYY117 pKa = 10.97LMQYY121 pKa = 9.69GGGWSSGVISLEE133 pKa = 3.71GLFNEE138 pKa = 3.81NRR140 pKa = 11.84LWRR143 pKa = 11.84NIWSKK148 pKa = 11.77SNDD151 pKa = 2.93GMDD154 pKa = 3.68LVRR157 pKa = 11.84YY158 pKa = 7.0FGCRR162 pKa = 11.84IRR164 pKa = 11.84LYY166 pKa = 8.47PTEE169 pKa = 4.0NQGYY173 pKa = 8.2LFWYY177 pKa = 8.41DD178 pKa = 3.52TEE180 pKa = 4.78FDD182 pKa = 3.61EE183 pKa = 5.1QQRR186 pKa = 11.84RR187 pKa = 11.84MLDD190 pKa = 3.49EE191 pKa = 3.97YY192 pKa = 9.6TQPSVMLQAKK202 pKa = 9.11NSRR205 pKa = 11.84LIVCKK210 pKa = 9.65QKK212 pKa = 9.55MPIRR216 pKa = 11.84RR217 pKa = 11.84RR218 pKa = 11.84VKK220 pKa = 10.4SIFIPPPAQLTTQWKK235 pKa = 8.5FQQEE239 pKa = 4.39LCQFPLFNWACICIDD254 pKa = 3.3MDD256 pKa = 4.61TPFDD260 pKa = 4.28YY261 pKa = 11.38NGAWRR266 pKa = 11.84NAWWLMRR273 pKa = 11.84RR274 pKa = 11.84LQNGNMEE281 pKa = 4.46YY282 pKa = 9.92IEE284 pKa = 4.12RR285 pKa = 11.84WGRR288 pKa = 11.84IPMTGDD294 pKa = 3.21TEE296 pKa = 4.91LPPADD301 pKa = 4.08DD302 pKa = 4.66FKK304 pKa = 11.68AGGVNKK310 pKa = 9.91NFKK313 pKa = 8.64PTGIQRR319 pKa = 11.84IYY321 pKa = 10.24PIVAVCLVEE330 pKa = 4.04GNKK333 pKa = 9.87RR334 pKa = 11.84VVKK337 pKa = 9.52WATVHH342 pKa = 6.44NGPIDD347 pKa = 3.43RR348 pKa = 11.84WRR350 pKa = 11.84KK351 pKa = 8.68KK352 pKa = 7.14QTGTLKK358 pKa = 10.77LSALRR363 pKa = 11.84RR364 pKa = 11.84LVLRR368 pKa = 11.84VCSEE372 pKa = 3.81SEE374 pKa = 4.12TYY376 pKa = 10.68YY377 pKa = 11.04KK378 pKa = 8.65WTASEE383 pKa = 4.09FTGAFQQDD391 pKa = 3.37WWPVSGTEE399 pKa = 4.79YY400 pKa = 10.41PLCTIKK406 pKa = 10.17MEE408 pKa = 4.57PEE410 pKa = 3.93FEE412 pKa = 4.24NPTVEE417 pKa = 4.01VWSWKK422 pKa = 8.95ATIPTAGTLKK432 pKa = 10.63DD433 pKa = 3.78YY434 pKa = 11.06FGLSSGQQWKK444 pKa = 8.76DD445 pKa = 2.72TDD447 pKa = 3.83FGRR450 pKa = 11.84LQLPRR455 pKa = 11.84SSHH458 pKa = 5.76NVDD461 pKa = 4.43FGHH464 pKa = 7.6KK465 pKa = 10.11ARR467 pKa = 11.84FGPFCVKK474 pKa = 10.27KK475 pKa = 10.35PPVEE479 pKa = 4.39FRR481 pKa = 11.84DD482 pKa = 3.9SAPNPLNIWVKK493 pKa = 8.38YY494 pKa = 7.74TFYY497 pKa = 10.76FQFGGMYY504 pKa = 9.72QPPTGIQDD512 pKa = 3.69PCTSNPTYY520 pKa = 9.73PVRR523 pKa = 11.84MVGAVTHH530 pKa = 6.51PKK532 pKa = 10.14YY533 pKa = 10.53AGQGGIATQIGDD545 pKa = 3.65QGITAASLRR554 pKa = 11.84AISAAPPNTYY564 pKa = 8.49TQSAFLKK571 pKa = 10.61APEE574 pKa = 4.24TEE576 pKa = 4.12KK577 pKa = 11.11EE578 pKa = 4.08EE579 pKa = 4.14EE580 pKa = 4.34RR581 pKa = 11.84EE582 pKa = 4.23SEE584 pKa = 4.25TSFTSAEE591 pKa = 4.16SSSEE595 pKa = 3.88GDD597 pKa = 3.7GSSDD601 pKa = 3.32DD602 pKa = 3.63QAEE605 pKa = 3.65RR606 pKa = 11.84RR607 pKa = 11.84AARR610 pKa = 11.84KK611 pKa = 9.09RR612 pKa = 11.84VIKK615 pKa = 10.69LLLKK619 pKa = 10.46RR620 pKa = 11.84LADD623 pKa = 3.62RR624 pKa = 11.84PVDD627 pKa = 3.43NKK629 pKa = 10.19RR630 pKa = 11.84RR631 pKa = 11.84RR632 pKa = 11.84FSEE635 pKa = 3.98
MM1 pKa = 7.79RR2 pKa = 11.84FRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84FGRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84YY14 pKa = 7.87YY15 pKa = 9.54RR16 pKa = 11.84KK17 pKa = 9.61RR18 pKa = 11.84RR19 pKa = 11.84GGWRR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84FRR27 pKa = 11.84IRR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84PWRR35 pKa = 11.84RR36 pKa = 11.84WRR38 pKa = 11.84VRR40 pKa = 11.84RR41 pKa = 11.84WRR43 pKa = 11.84RR44 pKa = 11.84SVFRR48 pKa = 11.84RR49 pKa = 11.84RR50 pKa = 11.84GRR52 pKa = 11.84RR53 pKa = 11.84ARR55 pKa = 11.84PYY57 pKa = 10.21RR58 pKa = 11.84ISAWNPKK65 pKa = 7.44VLRR68 pKa = 11.84NCRR71 pKa = 11.84ITGWWPVIQCMDD83 pKa = 2.9GMEE86 pKa = 4.32WIKK89 pKa = 10.41YY90 pKa = 9.99KK91 pKa = 10.93PMDD94 pKa = 3.88LRR96 pKa = 11.84VEE98 pKa = 4.09ANRR101 pKa = 11.84IFDD104 pKa = 3.72KK105 pKa = 10.88QGSKK109 pKa = 10.07IEE111 pKa = 4.16TEE113 pKa = 3.91QMGYY117 pKa = 10.97LMQYY121 pKa = 9.69GGGWSSGVISLEE133 pKa = 3.71GLFNEE138 pKa = 3.81NRR140 pKa = 11.84LWRR143 pKa = 11.84NIWSKK148 pKa = 11.77SNDD151 pKa = 2.93GMDD154 pKa = 3.68LVRR157 pKa = 11.84YY158 pKa = 7.0FGCRR162 pKa = 11.84IRR164 pKa = 11.84LYY166 pKa = 8.47PTEE169 pKa = 4.0NQGYY173 pKa = 8.2LFWYY177 pKa = 8.41DD178 pKa = 3.52TEE180 pKa = 4.78FDD182 pKa = 3.61EE183 pKa = 5.1QQRR186 pKa = 11.84RR187 pKa = 11.84MLDD190 pKa = 3.49EE191 pKa = 3.97YY192 pKa = 9.6TQPSVMLQAKK202 pKa = 9.11NSRR205 pKa = 11.84LIVCKK210 pKa = 9.65QKK212 pKa = 9.55MPIRR216 pKa = 11.84RR217 pKa = 11.84RR218 pKa = 11.84VKK220 pKa = 10.4SIFIPPPAQLTTQWKK235 pKa = 8.5FQQEE239 pKa = 4.39LCQFPLFNWACICIDD254 pKa = 3.3MDD256 pKa = 4.61TPFDD260 pKa = 4.28YY261 pKa = 11.38NGAWRR266 pKa = 11.84NAWWLMRR273 pKa = 11.84RR274 pKa = 11.84LQNGNMEE281 pKa = 4.46YY282 pKa = 9.92IEE284 pKa = 4.12RR285 pKa = 11.84WGRR288 pKa = 11.84IPMTGDD294 pKa = 3.21TEE296 pKa = 4.91LPPADD301 pKa = 4.08DD302 pKa = 4.66FKK304 pKa = 11.68AGGVNKK310 pKa = 9.91NFKK313 pKa = 8.64PTGIQRR319 pKa = 11.84IYY321 pKa = 10.24PIVAVCLVEE330 pKa = 4.04GNKK333 pKa = 9.87RR334 pKa = 11.84VVKK337 pKa = 9.52WATVHH342 pKa = 6.44NGPIDD347 pKa = 3.43RR348 pKa = 11.84WRR350 pKa = 11.84KK351 pKa = 8.68KK352 pKa = 7.14QTGTLKK358 pKa = 10.77LSALRR363 pKa = 11.84RR364 pKa = 11.84LVLRR368 pKa = 11.84VCSEE372 pKa = 3.81SEE374 pKa = 4.12TYY376 pKa = 10.68YY377 pKa = 11.04KK378 pKa = 8.65WTASEE383 pKa = 4.09FTGAFQQDD391 pKa = 3.37WWPVSGTEE399 pKa = 4.79YY400 pKa = 10.41PLCTIKK406 pKa = 10.17MEE408 pKa = 4.57PEE410 pKa = 3.93FEE412 pKa = 4.24NPTVEE417 pKa = 4.01VWSWKK422 pKa = 8.95ATIPTAGTLKK432 pKa = 10.63DD433 pKa = 3.78YY434 pKa = 11.06FGLSSGQQWKK444 pKa = 8.76DD445 pKa = 2.72TDD447 pKa = 3.83FGRR450 pKa = 11.84LQLPRR455 pKa = 11.84SSHH458 pKa = 5.76NVDD461 pKa = 4.43FGHH464 pKa = 7.6KK465 pKa = 10.11ARR467 pKa = 11.84FGPFCVKK474 pKa = 10.27KK475 pKa = 10.35PPVEE479 pKa = 4.39FRR481 pKa = 11.84DD482 pKa = 3.9SAPNPLNIWVKK493 pKa = 8.38YY494 pKa = 7.74TFYY497 pKa = 10.76FQFGGMYY504 pKa = 9.72QPPTGIQDD512 pKa = 3.69PCTSNPTYY520 pKa = 9.73PVRR523 pKa = 11.84MVGAVTHH530 pKa = 6.51PKK532 pKa = 10.14YY533 pKa = 10.53AGQGGIATQIGDD545 pKa = 3.65QGITAASLRR554 pKa = 11.84AISAAPPNTYY564 pKa = 8.49TQSAFLKK571 pKa = 10.61APEE574 pKa = 4.24TEE576 pKa = 4.12KK577 pKa = 11.11EE578 pKa = 4.08EE579 pKa = 4.14EE580 pKa = 4.34RR581 pKa = 11.84EE582 pKa = 4.23SEE584 pKa = 4.25TSFTSAEE591 pKa = 4.16SSSEE595 pKa = 3.88GDD597 pKa = 3.7GSSDD601 pKa = 3.32DD602 pKa = 3.63QAEE605 pKa = 3.65RR606 pKa = 11.84RR607 pKa = 11.84AARR610 pKa = 11.84KK611 pKa = 9.09RR612 pKa = 11.84VIKK615 pKa = 10.69LLLKK619 pKa = 10.46RR620 pKa = 11.84LADD623 pKa = 3.62RR624 pKa = 11.84PVDD627 pKa = 3.43NKK629 pKa = 10.19RR630 pKa = 11.84RR631 pKa = 11.84RR632 pKa = 11.84FSEE635 pKa = 3.98
Molecular weight: 74.96 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
932 |
73 |
635 |
310.7 |
36.09 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.223 ± 1.363 | 2.253 ± 0.676 |
6.76 ± 3.022 | 6.223 ± 0.917 |
4.077 ± 0.936 | 6.652 ± 1.233 |
1.502 ± 1.11 | 4.399 ± 0.582 |
5.794 ± 0.729 | 6.652 ± 1.21 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.253 ± 0.492 | 2.897 ± 1.215 |
6.545 ± 1.26 | 3.97 ± 1.145 |
10.408 ± 2.173 | 6.76 ± 1.842 |
5.579 ± 0.295 | 4.185 ± 0.858 |
3.97 ± 0.559 | 2.897 ± 1.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
