
Nesidiocoris tenuis virus
Taxonomy: Viruses; Riboviria; unclassified Riboviria
Average proteome isoelectric point is 7.39
Get precalculated fractions of proteins
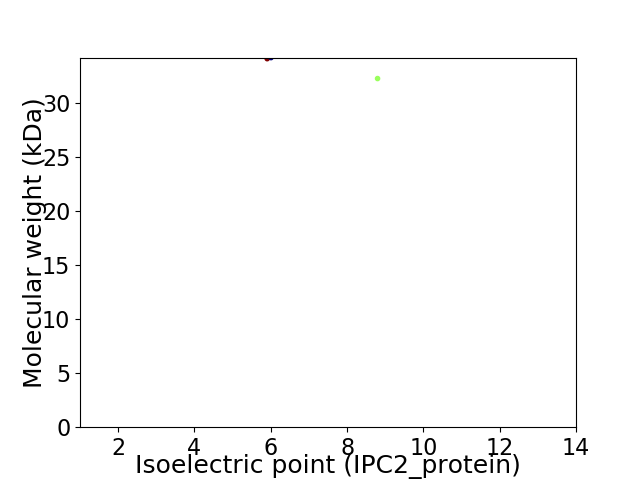
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
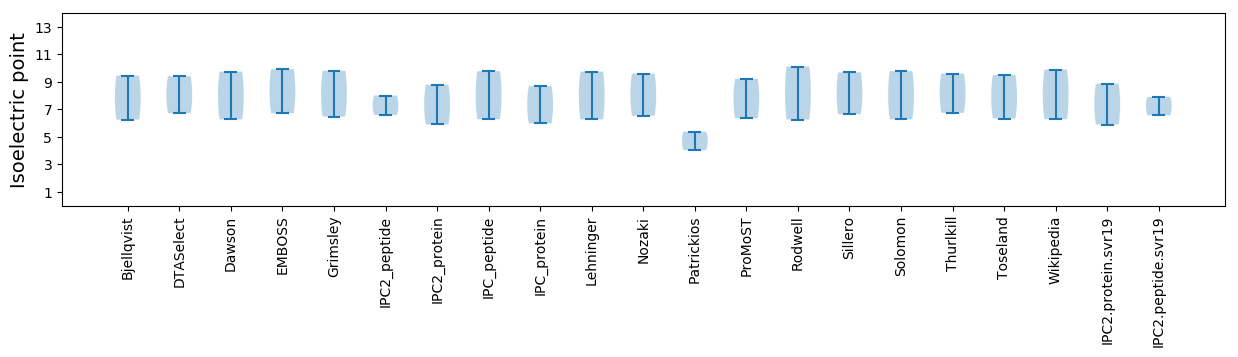
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L6YQ22|A0A1L6YQ22_9VIRU ORF2 OS=Nesidiocoris tenuis virus OX=1930921 PE=4 SV=1
MM1 pKa = 7.75ILPEE5 pKa = 4.66NPSSLYY11 pKa = 10.09EE12 pKa = 3.82WALNHH17 pKa = 5.86YY18 pKa = 10.68SYY20 pKa = 10.22MIKK23 pKa = 10.16SRR25 pKa = 11.84PKK27 pKa = 9.72PPLTEE32 pKa = 3.92EE33 pKa = 5.39AIDD36 pKa = 4.5EE37 pKa = 4.3YY38 pKa = 11.22TALQTIAYY46 pKa = 8.62HH47 pKa = 6.07EE48 pKa = 4.29KK49 pKa = 9.74FINAIFCPLMRR60 pKa = 11.84EE61 pKa = 4.2VKK63 pKa = 10.12KK64 pKa = 10.81RR65 pKa = 11.84LLAILDD71 pKa = 3.55PRR73 pKa = 11.84FVLFTDD79 pKa = 3.9MSPDD83 pKa = 3.56DD84 pKa = 3.65FADD87 pKa = 3.69RR88 pKa = 11.84LTRR91 pKa = 11.84TFSEE95 pKa = 4.26EE96 pKa = 4.36LFSKK100 pKa = 10.64DD101 pKa = 3.19YY102 pKa = 10.55VGHH105 pKa = 5.83VKK107 pKa = 10.56EE108 pKa = 3.85SDD110 pKa = 2.71IRR112 pKa = 11.84KK113 pKa = 9.16FDD115 pKa = 3.28KK116 pKa = 10.88SQQEE120 pKa = 3.93KK121 pKa = 10.39VLRR124 pKa = 11.84AEE126 pKa = 4.47QKK128 pKa = 10.31VLMLFGFPPQLAEE141 pKa = 3.54LWLRR145 pKa = 11.84VHH147 pKa = 7.19EE148 pKa = 4.15DD149 pKa = 3.18TILIDD154 pKa = 3.74RR155 pKa = 11.84GLGIMFRR162 pKa = 11.84VRR164 pKa = 11.84WHH166 pKa = 6.67RR167 pKa = 11.84KK168 pKa = 9.38SGDD171 pKa = 2.98ASTFLGNSLVLLMVLCSTYY190 pKa = 11.37DD191 pKa = 3.26LTKK194 pKa = 10.58AVMVVFPGDD203 pKa = 3.38DD204 pKa = 3.79FYY206 pKa = 11.04IIGPYY211 pKa = 10.09SLAIDD216 pKa = 3.92RR217 pKa = 11.84SAYY220 pKa = 9.85LAYY223 pKa = 10.47NYY225 pKa = 10.46NFEE228 pKa = 5.59SEE230 pKa = 4.28DD231 pKa = 3.52LDD233 pKa = 3.54YY234 pKa = 11.1KK235 pKa = 11.04YY236 pKa = 10.73IYY238 pKa = 9.77FCPKK242 pKa = 9.93FLPISIGGSVLGFLPTQLSMLSHH265 pKa = 7.48DD266 pKa = 3.74SLPRR270 pKa = 11.84SRR272 pKa = 11.84CSVNGQLDD280 pKa = 3.65QSAHH284 pKa = 5.73GARR287 pKa = 11.84CHH289 pKa = 4.88EE290 pKa = 4.29RR291 pKa = 11.84EE292 pKa = 3.96RR293 pKa = 11.84RR294 pKa = 3.57
MM1 pKa = 7.75ILPEE5 pKa = 4.66NPSSLYY11 pKa = 10.09EE12 pKa = 3.82WALNHH17 pKa = 5.86YY18 pKa = 10.68SYY20 pKa = 10.22MIKK23 pKa = 10.16SRR25 pKa = 11.84PKK27 pKa = 9.72PPLTEE32 pKa = 3.92EE33 pKa = 5.39AIDD36 pKa = 4.5EE37 pKa = 4.3YY38 pKa = 11.22TALQTIAYY46 pKa = 8.62HH47 pKa = 6.07EE48 pKa = 4.29KK49 pKa = 9.74FINAIFCPLMRR60 pKa = 11.84EE61 pKa = 4.2VKK63 pKa = 10.12KK64 pKa = 10.81RR65 pKa = 11.84LLAILDD71 pKa = 3.55PRR73 pKa = 11.84FVLFTDD79 pKa = 3.9MSPDD83 pKa = 3.56DD84 pKa = 3.65FADD87 pKa = 3.69RR88 pKa = 11.84LTRR91 pKa = 11.84TFSEE95 pKa = 4.26EE96 pKa = 4.36LFSKK100 pKa = 10.64DD101 pKa = 3.19YY102 pKa = 10.55VGHH105 pKa = 5.83VKK107 pKa = 10.56EE108 pKa = 3.85SDD110 pKa = 2.71IRR112 pKa = 11.84KK113 pKa = 9.16FDD115 pKa = 3.28KK116 pKa = 10.88SQQEE120 pKa = 3.93KK121 pKa = 10.39VLRR124 pKa = 11.84AEE126 pKa = 4.47QKK128 pKa = 10.31VLMLFGFPPQLAEE141 pKa = 3.54LWLRR145 pKa = 11.84VHH147 pKa = 7.19EE148 pKa = 4.15DD149 pKa = 3.18TILIDD154 pKa = 3.74RR155 pKa = 11.84GLGIMFRR162 pKa = 11.84VRR164 pKa = 11.84WHH166 pKa = 6.67RR167 pKa = 11.84KK168 pKa = 9.38SGDD171 pKa = 2.98ASTFLGNSLVLLMVLCSTYY190 pKa = 11.37DD191 pKa = 3.26LTKK194 pKa = 10.58AVMVVFPGDD203 pKa = 3.38DD204 pKa = 3.79FYY206 pKa = 11.04IIGPYY211 pKa = 10.09SLAIDD216 pKa = 3.92RR217 pKa = 11.84SAYY220 pKa = 9.85LAYY223 pKa = 10.47NYY225 pKa = 10.46NFEE228 pKa = 5.59SEE230 pKa = 4.28DD231 pKa = 3.52LDD233 pKa = 3.54YY234 pKa = 11.1KK235 pKa = 11.04YY236 pKa = 10.73IYY238 pKa = 9.77FCPKK242 pKa = 9.93FLPISIGGSVLGFLPTQLSMLSHH265 pKa = 7.48DD266 pKa = 3.74SLPRR270 pKa = 11.84SRR272 pKa = 11.84CSVNGQLDD280 pKa = 3.65QSAHH284 pKa = 5.73GARR287 pKa = 11.84CHH289 pKa = 4.88EE290 pKa = 4.29RR291 pKa = 11.84EE292 pKa = 3.96RR293 pKa = 11.84RR294 pKa = 3.57
Molecular weight: 34.08 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L6YQ22|A0A1L6YQ22_9VIRU ORF2 OS=Nesidiocoris tenuis virus OX=1930921 PE=4 SV=1
MM1 pKa = 7.6EE2 pKa = 5.45NLSVMYY8 pKa = 9.85QAVRR12 pKa = 11.84KK13 pKa = 10.0AMTIRR18 pKa = 11.84TPNSGKK24 pKa = 10.3LLNTEE29 pKa = 3.91TTRR32 pKa = 11.84LLRR35 pKa = 11.84SSNKK39 pKa = 10.23DD40 pKa = 3.07YY41 pKa = 11.54GLYY44 pKa = 10.14LVKK47 pKa = 10.15TGVFLLSPLARR58 pKa = 11.84DD59 pKa = 3.3HH60 pKa = 6.87RR61 pKa = 11.84FAYY64 pKa = 10.35DD65 pKa = 3.6GEE67 pKa = 4.24KK68 pKa = 10.42LVCFEE73 pKa = 4.94NKK75 pKa = 9.04MKK77 pKa = 9.86LTTTTNSDD85 pKa = 3.79YY86 pKa = 11.36LIVNCDD92 pKa = 3.49TEE94 pKa = 5.29RR95 pKa = 11.84MLEE98 pKa = 4.02RR99 pKa = 11.84QFYY102 pKa = 10.41SALKK106 pKa = 10.57NITLPTRR113 pKa = 11.84LPKK116 pKa = 10.4LRR118 pKa = 11.84FVLGVPGCGKK128 pKa = 8.65STYY131 pKa = 8.95IAKK134 pKa = 10.33ARR136 pKa = 11.84LPHH139 pKa = 5.93EE140 pKa = 4.65KK141 pKa = 10.1VWTATRR147 pKa = 11.84LGKK150 pKa = 10.18SDD152 pKa = 3.85VIKK155 pKa = 10.51KK156 pKa = 10.26IKK158 pKa = 9.8GDD160 pKa = 3.21KK161 pKa = 11.14DD162 pKa = 3.35MVRR165 pKa = 11.84TIGSILIKK173 pKa = 10.5GDD175 pKa = 3.05NTMTHH180 pKa = 6.36MKK182 pKa = 9.74NQPTRR187 pKa = 11.84VFVDD191 pKa = 3.92EE192 pKa = 5.22VIMAHH197 pKa = 6.42AGEE200 pKa = 4.5VMARR204 pKa = 11.84AQEE207 pKa = 3.95LDD209 pKa = 3.49IDD211 pKa = 4.13EE212 pKa = 5.05MICLGDD218 pKa = 3.99LKK220 pKa = 10.64QIPFIARR227 pKa = 11.84IAEE230 pKa = 4.29VKK232 pKa = 10.17LHH234 pKa = 5.47YY235 pKa = 10.85SKK237 pKa = 10.81ISDD240 pKa = 3.65MANKK244 pKa = 10.21DD245 pKa = 3.26IEE247 pKa = 4.42FLTNSHH253 pKa = 6.69RR254 pKa = 11.84VPADD258 pKa = 2.78IAVVLTRR265 pKa = 11.84YY266 pKa = 9.24FYY268 pKa = 11.31KK269 pKa = 10.86DD270 pKa = 3.19FVGNEE275 pKa = 4.1KK276 pKa = 10.26IKK278 pKa = 10.98LQTDD282 pKa = 3.37VV283 pKa = 3.79
MM1 pKa = 7.6EE2 pKa = 5.45NLSVMYY8 pKa = 9.85QAVRR12 pKa = 11.84KK13 pKa = 10.0AMTIRR18 pKa = 11.84TPNSGKK24 pKa = 10.3LLNTEE29 pKa = 3.91TTRR32 pKa = 11.84LLRR35 pKa = 11.84SSNKK39 pKa = 10.23DD40 pKa = 3.07YY41 pKa = 11.54GLYY44 pKa = 10.14LVKK47 pKa = 10.15TGVFLLSPLARR58 pKa = 11.84DD59 pKa = 3.3HH60 pKa = 6.87RR61 pKa = 11.84FAYY64 pKa = 10.35DD65 pKa = 3.6GEE67 pKa = 4.24KK68 pKa = 10.42LVCFEE73 pKa = 4.94NKK75 pKa = 9.04MKK77 pKa = 9.86LTTTTNSDD85 pKa = 3.79YY86 pKa = 11.36LIVNCDD92 pKa = 3.49TEE94 pKa = 5.29RR95 pKa = 11.84MLEE98 pKa = 4.02RR99 pKa = 11.84QFYY102 pKa = 10.41SALKK106 pKa = 10.57NITLPTRR113 pKa = 11.84LPKK116 pKa = 10.4LRR118 pKa = 11.84FVLGVPGCGKK128 pKa = 8.65STYY131 pKa = 8.95IAKK134 pKa = 10.33ARR136 pKa = 11.84LPHH139 pKa = 5.93EE140 pKa = 4.65KK141 pKa = 10.1VWTATRR147 pKa = 11.84LGKK150 pKa = 10.18SDD152 pKa = 3.85VIKK155 pKa = 10.51KK156 pKa = 10.26IKK158 pKa = 9.8GDD160 pKa = 3.21KK161 pKa = 11.14DD162 pKa = 3.35MVRR165 pKa = 11.84TIGSILIKK173 pKa = 10.5GDD175 pKa = 3.05NTMTHH180 pKa = 6.36MKK182 pKa = 9.74NQPTRR187 pKa = 11.84VFVDD191 pKa = 3.92EE192 pKa = 5.22VIMAHH197 pKa = 6.42AGEE200 pKa = 4.5VMARR204 pKa = 11.84AQEE207 pKa = 3.95LDD209 pKa = 3.49IDD211 pKa = 4.13EE212 pKa = 5.05MICLGDD218 pKa = 3.99LKK220 pKa = 10.64QIPFIARR227 pKa = 11.84IAEE230 pKa = 4.29VKK232 pKa = 10.17LHH234 pKa = 5.47YY235 pKa = 10.85SKK237 pKa = 10.81ISDD240 pKa = 3.65MANKK244 pKa = 10.21DD245 pKa = 3.26IEE247 pKa = 4.42FLTNSHH253 pKa = 6.69RR254 pKa = 11.84VPADD258 pKa = 2.78IAVVLTRR265 pKa = 11.84YY266 pKa = 9.24FYY268 pKa = 11.31KK269 pKa = 10.86DD270 pKa = 3.19FVGNEE275 pKa = 4.1KK276 pKa = 10.26IKK278 pKa = 10.98LQTDD282 pKa = 3.37VV283 pKa = 3.79
Molecular weight: 32.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
577 |
283 |
294 |
288.5 |
33.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.719 ± 0.185 | 1.56 ± 0.094 |
6.586 ± 0.145 | 5.546 ± 0.386 |
4.853 ± 0.85 | 4.679 ± 0.172 |
2.426 ± 0.197 | 6.239 ± 0.306 |
7.106 ± 1.341 | 11.265 ± 0.655 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.64 ± 0.387 | 3.466 ± 0.726 |
4.333 ± 0.742 | 2.426 ± 0.197 |
6.586 ± 0.145 | 6.412 ± 1.171 |
5.893 ± 1.439 | 6.239 ± 0.761 |
0.693 ± 0.219 | 4.333 ± 0.515 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
