
Grapevine vein clearing virus
Taxonomy: Viruses; Riboviria; Pararnavirae; Artverviricota; Revtraviricetes; Ortervirales; Caulimoviridae; Badnavirus
Average proteome isoelectric point is 6.88
Get precalculated fractions of proteins
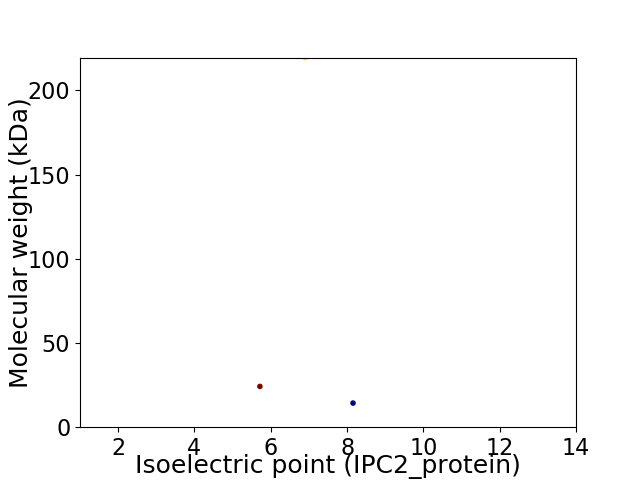
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F8TPA8|F8TPA8_9VIRU Uncharacterized protein OS=Grapevine vein clearing virus OX=1050407 PE=4 SV=1
MM1 pKa = 7.51QSIEE5 pKa = 4.04QQQFEE10 pKa = 4.57AEE12 pKa = 3.96IEE14 pKa = 3.92SWEE17 pKa = 4.07RR18 pKa = 11.84SEE20 pKa = 4.03RR21 pKa = 11.84TPLHH25 pKa = 6.78GYY27 pKa = 9.71RR28 pKa = 11.84DD29 pKa = 3.63LVEE32 pKa = 4.0YY33 pKa = 9.79PRR35 pKa = 11.84YY36 pKa = 9.69EE37 pKa = 4.2RR38 pKa = 11.84NQHH41 pKa = 5.38FPSAKK46 pKa = 8.29FPCYY50 pKa = 10.25HH51 pKa = 6.46FVAEE55 pKa = 4.62KK56 pKa = 11.07DD57 pKa = 3.88NVHH60 pKa = 5.63ATYY63 pKa = 10.08TKK65 pKa = 10.7GDD67 pKa = 4.73RR68 pKa = 11.84IPQLLNTLYY77 pKa = 10.7DD78 pKa = 3.72LQVNQCHH85 pKa = 5.34NQAVIYY91 pKa = 10.46DD92 pKa = 4.59RR93 pKa = 11.84IQLLSRR99 pKa = 11.84YY100 pKa = 6.67TVRR103 pKa = 11.84KK104 pKa = 9.27GKK106 pKa = 9.01PLPAIPEE113 pKa = 3.99EE114 pKa = 4.51SVLKK118 pKa = 10.49EE119 pKa = 4.0PEE121 pKa = 3.99EE122 pKa = 4.37SSTEE126 pKa = 3.86LKK128 pKa = 10.51HH129 pKa = 7.36QIEE132 pKa = 4.77LLRR135 pKa = 11.84ADD137 pKa = 3.85LRR139 pKa = 11.84EE140 pKa = 3.9IKK142 pKa = 10.56ANQSSLRR149 pKa = 11.84LAISEE154 pKa = 3.86IRR156 pKa = 11.84EE157 pKa = 4.35SITDD161 pKa = 3.67LTARR165 pKa = 11.84EE166 pKa = 4.27SAPKK170 pKa = 9.86PIEE173 pKa = 4.39AEE175 pKa = 4.06TAYY178 pKa = 9.53LTAQLKK184 pKa = 9.62VQVQEE189 pKa = 3.9IKK191 pKa = 9.77TALTEE196 pKa = 3.92IKK198 pKa = 9.56TFARR202 pKa = 11.84TLVPEE207 pKa = 4.46RR208 pKa = 4.22
MM1 pKa = 7.51QSIEE5 pKa = 4.04QQQFEE10 pKa = 4.57AEE12 pKa = 3.96IEE14 pKa = 3.92SWEE17 pKa = 4.07RR18 pKa = 11.84SEE20 pKa = 4.03RR21 pKa = 11.84TPLHH25 pKa = 6.78GYY27 pKa = 9.71RR28 pKa = 11.84DD29 pKa = 3.63LVEE32 pKa = 4.0YY33 pKa = 9.79PRR35 pKa = 11.84YY36 pKa = 9.69EE37 pKa = 4.2RR38 pKa = 11.84NQHH41 pKa = 5.38FPSAKK46 pKa = 8.29FPCYY50 pKa = 10.25HH51 pKa = 6.46FVAEE55 pKa = 4.62KK56 pKa = 11.07DD57 pKa = 3.88NVHH60 pKa = 5.63ATYY63 pKa = 10.08TKK65 pKa = 10.7GDD67 pKa = 4.73RR68 pKa = 11.84IPQLLNTLYY77 pKa = 10.7DD78 pKa = 3.72LQVNQCHH85 pKa = 5.34NQAVIYY91 pKa = 10.46DD92 pKa = 4.59RR93 pKa = 11.84IQLLSRR99 pKa = 11.84YY100 pKa = 6.67TVRR103 pKa = 11.84KK104 pKa = 9.27GKK106 pKa = 9.01PLPAIPEE113 pKa = 3.99EE114 pKa = 4.51SVLKK118 pKa = 10.49EE119 pKa = 4.0PEE121 pKa = 3.99EE122 pKa = 4.37SSTEE126 pKa = 3.86LKK128 pKa = 10.51HH129 pKa = 7.36QIEE132 pKa = 4.77LLRR135 pKa = 11.84ADD137 pKa = 3.85LRR139 pKa = 11.84EE140 pKa = 3.9IKK142 pKa = 10.56ANQSSLRR149 pKa = 11.84LAISEE154 pKa = 3.86IRR156 pKa = 11.84EE157 pKa = 4.35SITDD161 pKa = 3.67LTARR165 pKa = 11.84EE166 pKa = 4.27SAPKK170 pKa = 9.86PIEE173 pKa = 4.39AEE175 pKa = 4.06TAYY178 pKa = 9.53LTAQLKK184 pKa = 9.62VQVQEE189 pKa = 3.9IKK191 pKa = 9.77TALTEE196 pKa = 3.92IKK198 pKa = 9.56TFARR202 pKa = 11.84TLVPEE207 pKa = 4.46RR208 pKa = 4.22
Molecular weight: 24.22 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F8TPA9|F8TPA9_9VIRU Reverse transcriptase OS=Grapevine vein clearing virus OX=1050407 PE=4 SV=2
MM1 pKa = 7.02STWQIAAAAEE11 pKa = 4.31EE12 pKa = 4.38YY13 pKa = 10.94KK14 pKa = 11.01NAIKK18 pKa = 9.56ATATLTKK25 pKa = 10.32DD26 pKa = 2.95EE27 pKa = 4.37RR28 pKa = 11.84AVGFVKK34 pKa = 10.35PHH36 pKa = 5.19EE37 pKa = 4.38FEE39 pKa = 5.42PNFSDD44 pKa = 5.0TNIQRR49 pKa = 11.84QNNTLIHH56 pKa = 6.6LLIQSLEE63 pKa = 4.2EE64 pKa = 4.01IKK66 pKa = 10.55EE67 pKa = 3.81LRR69 pKa = 11.84AQVQTLNDD77 pKa = 4.19RR78 pKa = 11.84IITLEE83 pKa = 3.92KK84 pKa = 10.39GKK86 pKa = 10.86APVTLPDD93 pKa = 3.54NVVEE97 pKa = 4.78QISTQLKK104 pKa = 7.93EE105 pKa = 4.4AKK107 pKa = 9.71FGQPKK112 pKa = 9.87EE113 pKa = 4.06GLVKK117 pKa = 9.62GTKK120 pKa = 8.65GTFRR124 pKa = 11.84VWKK127 pKa = 10.13
MM1 pKa = 7.02STWQIAAAAEE11 pKa = 4.31EE12 pKa = 4.38YY13 pKa = 10.94KK14 pKa = 11.01NAIKK18 pKa = 9.56ATATLTKK25 pKa = 10.32DD26 pKa = 2.95EE27 pKa = 4.37RR28 pKa = 11.84AVGFVKK34 pKa = 10.35PHH36 pKa = 5.19EE37 pKa = 4.38FEE39 pKa = 5.42PNFSDD44 pKa = 5.0TNIQRR49 pKa = 11.84QNNTLIHH56 pKa = 6.6LLIQSLEE63 pKa = 4.2EE64 pKa = 4.01IKK66 pKa = 10.55EE67 pKa = 3.81LRR69 pKa = 11.84AQVQTLNDD77 pKa = 4.19RR78 pKa = 11.84IITLEE83 pKa = 3.92KK84 pKa = 10.39GKK86 pKa = 10.86APVTLPDD93 pKa = 3.54NVVEE97 pKa = 4.78QISTQLKK104 pKa = 7.93EE105 pKa = 4.4AKK107 pKa = 9.71FGQPKK112 pKa = 9.87EE113 pKa = 4.06GLVKK117 pKa = 9.62GTKK120 pKa = 8.65GTFRR124 pKa = 11.84VWKK127 pKa = 10.13
Molecular weight: 14.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2276 |
127 |
1941 |
758.7 |
86.01 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.854 ± 0.608 | 1.538 ± 0.484 |
5.141 ± 0.901 | 7.645 ± 1.671 |
3.032 ± 0.229 | 5.8 ± 1.561 |
2.417 ± 0.197 | 6.634 ± 0.126 |
6.591 ± 0.808 | 8.172 ± 0.691 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.197 ± 0.774 | 3.515 ± 0.439 |
5.624 ± 0.391 | 5.844 ± 0.639 |
6.195 ± 0.56 | 6.591 ± 0.888 |
6.151 ± 0.897 | 5.316 ± 0.226 |
1.406 ± 0.289 | 3.339 ± 0.564 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
