
Capybara microvirus Cap1_SP_168
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.17
Get precalculated fractions of proteins
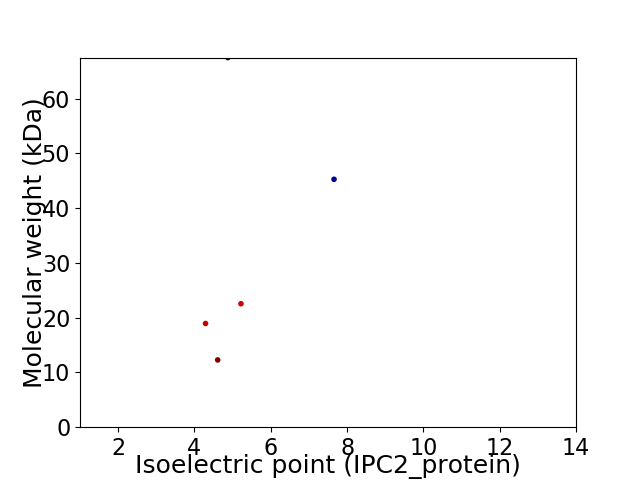
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W821|A0A4P8W821_9VIRU Minor capsid protein OS=Capybara microvirus Cap1_SP_168 OX=2585403 PE=4 SV=1
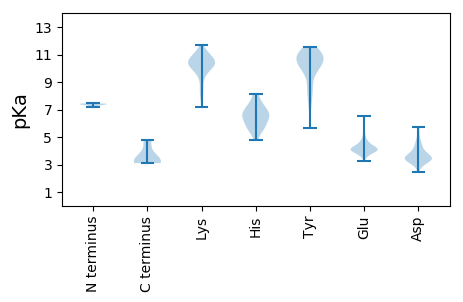
MM1 pKa = 7.39KK2 pKa = 10.24FSSRR6 pKa = 11.84YY7 pKa = 5.65EE8 pKa = 4.07HH9 pKa = 7.18PVTPITNPGTEE20 pKa = 3.79FRR22 pKa = 11.84NDD24 pKa = 3.28YY25 pKa = 8.8QAHH28 pKa = 5.7YY29 pKa = 10.6DD30 pKa = 3.67EE31 pKa = 6.54DD32 pKa = 4.43GVLVTVKK39 pKa = 10.62VGEE42 pKa = 4.19TNIYY46 pKa = 11.04QMIQSYY52 pKa = 10.87ADD54 pKa = 3.32SVDD57 pKa = 2.93IHH59 pKa = 7.54ILLEE63 pKa = 4.2QFGSGYY69 pKa = 11.14AEE71 pKa = 3.71ALNRR75 pKa = 11.84FEE77 pKa = 5.95GFYY80 pKa = 11.08ADD82 pKa = 4.79ISEE85 pKa = 5.3LPDD88 pKa = 3.46NYY90 pKa = 10.74VGMLNTIEE98 pKa = 4.25TTRR101 pKa = 11.84SYY103 pKa = 11.21FDD105 pKa = 4.57KK106 pKa = 10.94LPADD110 pKa = 3.38IRR112 pKa = 11.84SKK114 pKa = 10.95FNNDD118 pKa = 2.46FSQFLVASQQPDD130 pKa = 3.62FLSNFAPAVPISNMYY145 pKa = 9.98VPGEE149 pKa = 4.05NVPGEE154 pKa = 4.21NVPGEE159 pKa = 4.16NVPGEE164 pKa = 4.42KK165 pKa = 10.18EE166 pKa = 4.0NQQ168 pKa = 3.16
MM1 pKa = 7.39KK2 pKa = 10.24FSSRR6 pKa = 11.84YY7 pKa = 5.65EE8 pKa = 4.07HH9 pKa = 7.18PVTPITNPGTEE20 pKa = 3.79FRR22 pKa = 11.84NDD24 pKa = 3.28YY25 pKa = 8.8QAHH28 pKa = 5.7YY29 pKa = 10.6DD30 pKa = 3.67EE31 pKa = 6.54DD32 pKa = 4.43GVLVTVKK39 pKa = 10.62VGEE42 pKa = 4.19TNIYY46 pKa = 11.04QMIQSYY52 pKa = 10.87ADD54 pKa = 3.32SVDD57 pKa = 2.93IHH59 pKa = 7.54ILLEE63 pKa = 4.2QFGSGYY69 pKa = 11.14AEE71 pKa = 3.71ALNRR75 pKa = 11.84FEE77 pKa = 5.95GFYY80 pKa = 11.08ADD82 pKa = 4.79ISEE85 pKa = 5.3LPDD88 pKa = 3.46NYY90 pKa = 10.74VGMLNTIEE98 pKa = 4.25TTRR101 pKa = 11.84SYY103 pKa = 11.21FDD105 pKa = 4.57KK106 pKa = 10.94LPADD110 pKa = 3.38IRR112 pKa = 11.84SKK114 pKa = 10.95FNNDD118 pKa = 2.46FSQFLVASQQPDD130 pKa = 3.62FLSNFAPAVPISNMYY145 pKa = 9.98VPGEE149 pKa = 4.05NVPGEE154 pKa = 4.21NVPGEE159 pKa = 4.16NVPGEE164 pKa = 4.42KK165 pKa = 10.18EE166 pKa = 4.0NQQ168 pKa = 3.16
Molecular weight: 18.93 kDa
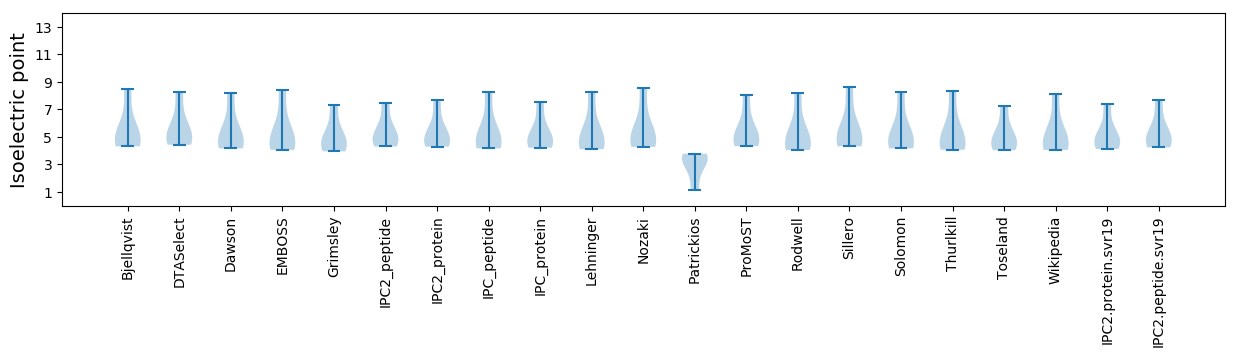
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W5K6|A0A4P8W5K6_9VIRU Minor capsid protein OS=Capybara microvirus Cap1_SP_168 OX=2585403 PE=4 SV=1
MM1 pKa = 7.18TCYY4 pKa = 10.43HH5 pKa = 6.5PLKK8 pKa = 10.64RR9 pKa = 11.84FTTGFNPEE17 pKa = 3.91TGKK20 pKa = 11.07SIGKK24 pKa = 8.06VVPYY28 pKa = 10.39RR29 pKa = 11.84VDD31 pKa = 3.24HH32 pKa = 6.14LEE34 pKa = 3.64KK35 pKa = 10.43RR36 pKa = 11.84YY37 pKa = 10.7GDD39 pKa = 3.37DD40 pKa = 4.05RR41 pKa = 11.84FYY43 pKa = 11.3QVYY46 pKa = 10.76DD47 pKa = 3.44PVDD50 pKa = 3.46YY51 pKa = 7.82TTYY54 pKa = 10.17TINGTKK60 pKa = 9.97FEE62 pKa = 4.31NMPLKK67 pKa = 10.61PSDD70 pKa = 3.2VRR72 pKa = 11.84IYY74 pKa = 10.96SDD76 pKa = 4.3YY77 pKa = 11.44EE78 pKa = 4.05VIPCGKK84 pKa = 10.37CIGCRR89 pKa = 11.84LDD91 pKa = 3.48YY92 pKa = 11.29SRR94 pKa = 11.84MWSSRR99 pKa = 11.84LMAEE103 pKa = 4.63LPYY106 pKa = 10.6HH107 pKa = 6.1HH108 pKa = 6.73EE109 pKa = 3.91AWFLTLTYY117 pKa = 10.95NDD119 pKa = 4.11LCLPSHH125 pKa = 5.86IVEE128 pKa = 4.24RR129 pKa = 11.84DD130 pKa = 3.21GKK132 pKa = 10.67LVEE135 pKa = 4.14THH137 pKa = 6.52SLVKK141 pKa = 10.55KK142 pKa = 10.05HH143 pKa = 6.02FQDD146 pKa = 3.16FMKK149 pKa = 10.42RR150 pKa = 11.84LRR152 pKa = 11.84RR153 pKa = 11.84RR154 pKa = 11.84FPDD157 pKa = 3.66GSIKK161 pKa = 10.52YY162 pKa = 7.41YY163 pKa = 10.8CCGEE167 pKa = 4.16YY168 pKa = 10.47GSKK171 pKa = 10.46SMRR174 pKa = 11.84PHH176 pKa = 4.77YY177 pKa = 10.13HH178 pKa = 6.99AILFFDD184 pKa = 5.04SSLQDD189 pKa = 3.29YY190 pKa = 8.01VFPNGVINTNKK201 pKa = 9.44LHH203 pKa = 6.41KK204 pKa = 10.38VQDD207 pKa = 3.81GYY209 pKa = 11.32RR210 pKa = 11.84YY211 pKa = 9.64YY212 pKa = 11.36VNRR215 pKa = 11.84LIDD218 pKa = 3.71QLWYY222 pKa = 11.07VSDD225 pKa = 4.71CPDD228 pKa = 2.94VSAGFHH234 pKa = 6.45YY235 pKa = 9.28LTQVTYY241 pKa = 8.59EE242 pKa = 4.16TICYY246 pKa = 6.19VTQYY250 pKa = 8.28VTKK253 pKa = 10.41KK254 pKa = 10.55LNGDD258 pKa = 3.47AASIYY263 pKa = 9.84QEE265 pKa = 3.94YY266 pKa = 10.72DD267 pKa = 3.25LEE269 pKa = 4.8PEE271 pKa = 4.26FSLISTKK278 pKa = 10.19PALGRR283 pKa = 11.84QFYY286 pKa = 9.58EE287 pKa = 3.68EE288 pKa = 4.44HH289 pKa = 6.54KK290 pKa = 11.16AEE292 pKa = 4.66IYY294 pKa = 11.04ADD296 pKa = 3.47DD297 pKa = 4.18KK298 pKa = 11.52FVFEE302 pKa = 5.11SNGKK306 pKa = 8.5PVIFRR311 pKa = 11.84PPAYY315 pKa = 9.65YY316 pKa = 10.63DD317 pKa = 3.21HH318 pKa = 7.17LFDD321 pKa = 5.32IEE323 pKa = 4.3APEE326 pKa = 4.38IMSQKK331 pKa = 9.57KK332 pKa = 10.06DD333 pKa = 3.3KK334 pKa = 10.89RR335 pKa = 11.84KK336 pKa = 9.6EE337 pKa = 3.49LAINHH342 pKa = 5.44VDD344 pKa = 3.16ALLRR348 pKa = 11.84QTDD351 pKa = 3.28KK352 pKa = 11.23DD353 pKa = 4.06YY354 pKa = 10.93PAILADD360 pKa = 3.5QEE362 pKa = 4.77SAFKK366 pKa = 10.78SRR368 pKa = 11.84ASSRR372 pKa = 11.84MKK374 pKa = 10.68AKK376 pKa = 10.32YY377 pKa = 9.92NKK379 pKa = 10.08AKK381 pKa = 9.9GRR383 pKa = 11.84RR384 pKa = 11.84VV385 pKa = 3.1
MM1 pKa = 7.18TCYY4 pKa = 10.43HH5 pKa = 6.5PLKK8 pKa = 10.64RR9 pKa = 11.84FTTGFNPEE17 pKa = 3.91TGKK20 pKa = 11.07SIGKK24 pKa = 8.06VVPYY28 pKa = 10.39RR29 pKa = 11.84VDD31 pKa = 3.24HH32 pKa = 6.14LEE34 pKa = 3.64KK35 pKa = 10.43RR36 pKa = 11.84YY37 pKa = 10.7GDD39 pKa = 3.37DD40 pKa = 4.05RR41 pKa = 11.84FYY43 pKa = 11.3QVYY46 pKa = 10.76DD47 pKa = 3.44PVDD50 pKa = 3.46YY51 pKa = 7.82TTYY54 pKa = 10.17TINGTKK60 pKa = 9.97FEE62 pKa = 4.31NMPLKK67 pKa = 10.61PSDD70 pKa = 3.2VRR72 pKa = 11.84IYY74 pKa = 10.96SDD76 pKa = 4.3YY77 pKa = 11.44EE78 pKa = 4.05VIPCGKK84 pKa = 10.37CIGCRR89 pKa = 11.84LDD91 pKa = 3.48YY92 pKa = 11.29SRR94 pKa = 11.84MWSSRR99 pKa = 11.84LMAEE103 pKa = 4.63LPYY106 pKa = 10.6HH107 pKa = 6.1HH108 pKa = 6.73EE109 pKa = 3.91AWFLTLTYY117 pKa = 10.95NDD119 pKa = 4.11LCLPSHH125 pKa = 5.86IVEE128 pKa = 4.24RR129 pKa = 11.84DD130 pKa = 3.21GKK132 pKa = 10.67LVEE135 pKa = 4.14THH137 pKa = 6.52SLVKK141 pKa = 10.55KK142 pKa = 10.05HH143 pKa = 6.02FQDD146 pKa = 3.16FMKK149 pKa = 10.42RR150 pKa = 11.84LRR152 pKa = 11.84RR153 pKa = 11.84RR154 pKa = 11.84FPDD157 pKa = 3.66GSIKK161 pKa = 10.52YY162 pKa = 7.41YY163 pKa = 10.8CCGEE167 pKa = 4.16YY168 pKa = 10.47GSKK171 pKa = 10.46SMRR174 pKa = 11.84PHH176 pKa = 4.77YY177 pKa = 10.13HH178 pKa = 6.99AILFFDD184 pKa = 5.04SSLQDD189 pKa = 3.29YY190 pKa = 8.01VFPNGVINTNKK201 pKa = 9.44LHH203 pKa = 6.41KK204 pKa = 10.38VQDD207 pKa = 3.81GYY209 pKa = 11.32RR210 pKa = 11.84YY211 pKa = 9.64YY212 pKa = 11.36VNRR215 pKa = 11.84LIDD218 pKa = 3.71QLWYY222 pKa = 11.07VSDD225 pKa = 4.71CPDD228 pKa = 2.94VSAGFHH234 pKa = 6.45YY235 pKa = 9.28LTQVTYY241 pKa = 8.59EE242 pKa = 4.16TICYY246 pKa = 6.19VTQYY250 pKa = 8.28VTKK253 pKa = 10.41KK254 pKa = 10.55LNGDD258 pKa = 3.47AASIYY263 pKa = 9.84QEE265 pKa = 3.94YY266 pKa = 10.72DD267 pKa = 3.25LEE269 pKa = 4.8PEE271 pKa = 4.26FSLISTKK278 pKa = 10.19PALGRR283 pKa = 11.84QFYY286 pKa = 9.58EE287 pKa = 3.68EE288 pKa = 4.44HH289 pKa = 6.54KK290 pKa = 11.16AEE292 pKa = 4.66IYY294 pKa = 11.04ADD296 pKa = 3.47DD297 pKa = 4.18KK298 pKa = 11.52FVFEE302 pKa = 5.11SNGKK306 pKa = 8.5PVIFRR311 pKa = 11.84PPAYY315 pKa = 9.65YY316 pKa = 10.63DD317 pKa = 3.21HH318 pKa = 7.17LFDD321 pKa = 5.32IEE323 pKa = 4.3APEE326 pKa = 4.38IMSQKK331 pKa = 9.57KK332 pKa = 10.06DD333 pKa = 3.3KK334 pKa = 10.89RR335 pKa = 11.84KK336 pKa = 9.6EE337 pKa = 3.49LAINHH342 pKa = 5.44VDD344 pKa = 3.16ALLRR348 pKa = 11.84QTDD351 pKa = 3.28KK352 pKa = 11.23DD353 pKa = 4.06YY354 pKa = 10.93PAILADD360 pKa = 3.5QEE362 pKa = 4.77SAFKK366 pKa = 10.78SRR368 pKa = 11.84ASSRR372 pKa = 11.84MKK374 pKa = 10.68AKK376 pKa = 10.32YY377 pKa = 9.92NKK379 pKa = 10.08AKK381 pKa = 9.9GRR383 pKa = 11.84RR384 pKa = 11.84VV385 pKa = 3.1
Molecular weight: 45.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1479 |
107 |
598 |
295.8 |
33.3 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.384 ± 2.346 | 1.014 ± 0.42 |
6.829 ± 0.624 | 5.477 ± 0.694 |
5.341 ± 0.369 | 6.558 ± 0.775 |
2.366 ± 0.392 | 5.341 ± 0.477 |
5.139 ± 0.907 | 6.694 ± 0.471 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.164 ± 0.105 | 5.274 ± 1.038 |
5.003 ± 0.732 | 3.854 ± 0.38 |
4.124 ± 0.736 | 7.911 ± 0.99 |
5.274 ± 0.198 | 5.68 ± 0.77 |
0.947 ± 0.184 | 6.626 ± 0.586 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
