
Walleye dermal sarcoma virus (WDSV)
Taxonomy: Viruses; Riboviria; Pararnavirae; Artverviricota; Revtraviricetes; Ortervirales; Retroviridae; Orthoretrovirinae; Epsilonretrovirus
Average proteome isoelectric point is 7.08
Get precalculated fractions of proteins
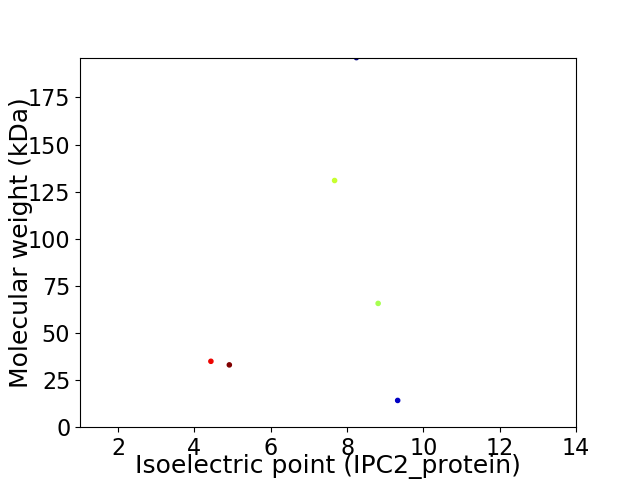
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q88940|ORFB_WDSV ORF-B protein OS=Walleye dermal sarcoma virus OX=39720 PE=1 SV=1
MM1 pKa = 7.42FSDD4 pKa = 4.06SDD6 pKa = 3.96SSDD9 pKa = 3.57EE10 pKa = 3.95EE11 pKa = 3.94LSRR14 pKa = 11.84IITDD18 pKa = 3.06IDD20 pKa = 3.84EE21 pKa = 4.57SPQDD25 pKa = 3.56IQQSLHH31 pKa = 5.68LRR33 pKa = 11.84AGVSPEE39 pKa = 3.48ARR41 pKa = 11.84VPPGGLTQEE50 pKa = 4.37EE51 pKa = 4.51WTIDD55 pKa = 3.64YY56 pKa = 9.99FSTYY60 pKa = 8.21HH61 pKa = 6.24TPLLNPGIPHH71 pKa = 7.5EE72 pKa = 4.07LTQRR76 pKa = 11.84CTDD79 pKa = 3.45YY80 pKa = 10.12TASILRR86 pKa = 11.84RR87 pKa = 11.84ASAKK91 pKa = 9.57MIQWDD96 pKa = 4.05YY97 pKa = 11.76VFYY100 pKa = 10.89LLPRR104 pKa = 11.84VWIMFPFIARR114 pKa = 11.84EE115 pKa = 4.08GLSHH119 pKa = 6.85LTHH122 pKa = 7.41LLTLTTSVLSATSLVFGWDD141 pKa = 3.09LTVIEE146 pKa = 5.52LCNEE150 pKa = 3.77MNIQGVYY157 pKa = 9.81LPEE160 pKa = 4.47VIEE163 pKa = 4.14WLAQFSFLFTHH174 pKa = 5.57VTLIVVSDD182 pKa = 3.26GMMDD186 pKa = 4.72LLLMFPMDD194 pKa = 4.42IEE196 pKa = 4.23EE197 pKa = 4.3QPLAINIALHH207 pKa = 6.64ALQTSYY213 pKa = 11.27TIMTPILFASPLLRR227 pKa = 11.84IISCVLYY234 pKa = 10.83ACGHH238 pKa = 6.1CPSARR243 pKa = 11.84MLYY246 pKa = 10.26AYY248 pKa = 9.41TIMNRR253 pKa = 11.84YY254 pKa = 6.85TGEE257 pKa = 4.47SIAEE261 pKa = 3.82MHH263 pKa = 6.17TGFRR267 pKa = 11.84CFRR270 pKa = 11.84DD271 pKa = 3.35QMIAYY276 pKa = 10.07DD277 pKa = 3.7MEE279 pKa = 4.33FTNFLRR285 pKa = 11.84DD286 pKa = 3.54LTEE289 pKa = 3.94EE290 pKa = 4.04EE291 pKa = 4.74TPVLEE296 pKa = 4.11ITEE299 pKa = 4.59PEE301 pKa = 4.14PSPTEE306 pKa = 3.71
MM1 pKa = 7.42FSDD4 pKa = 4.06SDD6 pKa = 3.96SSDD9 pKa = 3.57EE10 pKa = 3.95EE11 pKa = 3.94LSRR14 pKa = 11.84IITDD18 pKa = 3.06IDD20 pKa = 3.84EE21 pKa = 4.57SPQDD25 pKa = 3.56IQQSLHH31 pKa = 5.68LRR33 pKa = 11.84AGVSPEE39 pKa = 3.48ARR41 pKa = 11.84VPPGGLTQEE50 pKa = 4.37EE51 pKa = 4.51WTIDD55 pKa = 3.64YY56 pKa = 9.99FSTYY60 pKa = 8.21HH61 pKa = 6.24TPLLNPGIPHH71 pKa = 7.5EE72 pKa = 4.07LTQRR76 pKa = 11.84CTDD79 pKa = 3.45YY80 pKa = 10.12TASILRR86 pKa = 11.84RR87 pKa = 11.84ASAKK91 pKa = 9.57MIQWDD96 pKa = 4.05YY97 pKa = 11.76VFYY100 pKa = 10.89LLPRR104 pKa = 11.84VWIMFPFIARR114 pKa = 11.84EE115 pKa = 4.08GLSHH119 pKa = 6.85LTHH122 pKa = 7.41LLTLTTSVLSATSLVFGWDD141 pKa = 3.09LTVIEE146 pKa = 5.52LCNEE150 pKa = 3.77MNIQGVYY157 pKa = 9.81LPEE160 pKa = 4.47VIEE163 pKa = 4.14WLAQFSFLFTHH174 pKa = 5.57VTLIVVSDD182 pKa = 3.26GMMDD186 pKa = 4.72LLLMFPMDD194 pKa = 4.42IEE196 pKa = 4.23EE197 pKa = 4.3QPLAINIALHH207 pKa = 6.64ALQTSYY213 pKa = 11.27TIMTPILFASPLLRR227 pKa = 11.84IISCVLYY234 pKa = 10.83ACGHH238 pKa = 6.1CPSARR243 pKa = 11.84MLYY246 pKa = 10.26AYY248 pKa = 9.41TIMNRR253 pKa = 11.84YY254 pKa = 6.85TGEE257 pKa = 4.47SIAEE261 pKa = 3.82MHH263 pKa = 6.17TGFRR267 pKa = 11.84CFRR270 pKa = 11.84DD271 pKa = 3.35QMIAYY276 pKa = 10.07DD277 pKa = 3.7MEE279 pKa = 4.33FTNFLRR285 pKa = 11.84DD286 pKa = 3.54LTEE289 pKa = 3.94EE290 pKa = 4.04EE291 pKa = 4.74TPVLEE296 pKa = 4.11ITEE299 pKa = 4.59PEE301 pKa = 4.14PSPTEE306 pKa = 3.71
Molecular weight: 34.97 kDa
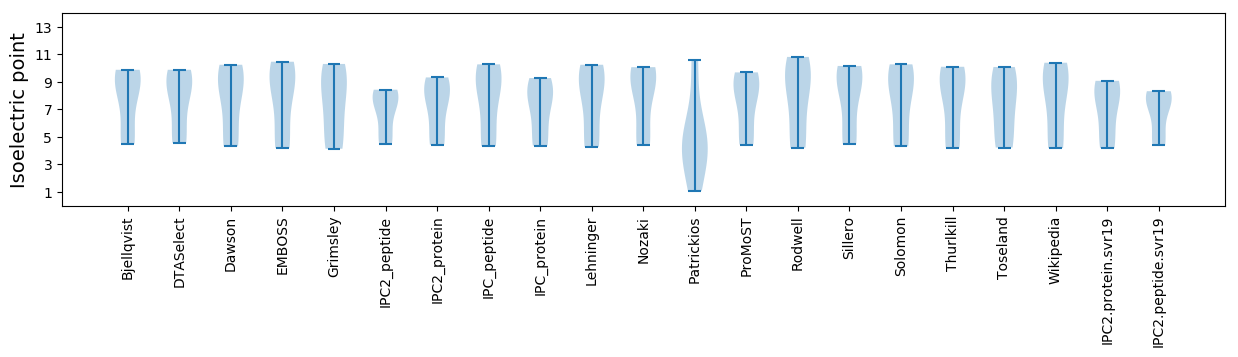
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q88937|GAG_WDSV Gag polyprotein OS=Walleye dermal sarcoma virus OX=39720 GN=gag PE=1 SV=1
MM1 pKa = 7.41AWYY4 pKa = 8.86HH5 pKa = 4.09QHH7 pKa = 6.47RR8 pKa = 11.84WHH10 pKa = 7.21LDD12 pKa = 3.19YY13 pKa = 11.11SIPRR17 pKa = 11.84QNLQAFLTTITFIDD31 pKa = 4.01PQFKK35 pKa = 10.05IQEE38 pKa = 4.14NGLTEE43 pKa = 4.6GEE45 pKa = 4.44YY46 pKa = 9.08KK47 pKa = 9.1TQIVKK52 pKa = 10.34QIIPQLCRR60 pKa = 11.84IPNQNSPPPIWVQGPRR76 pKa = 11.84IKK78 pKa = 10.85GDD80 pKa = 3.67PTWLKK85 pKa = 10.71INAKK89 pKa = 10.0FITEE93 pKa = 5.42LIPKK97 pKa = 9.79QKK99 pKa = 8.72GTKK102 pKa = 9.81NISTKK107 pKa = 10.15TYY109 pKa = 9.75LSRR112 pKa = 11.84LFVIWLQNN120 pKa = 3.18
MM1 pKa = 7.41AWYY4 pKa = 8.86HH5 pKa = 4.09QHH7 pKa = 6.47RR8 pKa = 11.84WHH10 pKa = 7.21LDD12 pKa = 3.19YY13 pKa = 11.11SIPRR17 pKa = 11.84QNLQAFLTTITFIDD31 pKa = 4.01PQFKK35 pKa = 10.05IQEE38 pKa = 4.14NGLTEE43 pKa = 4.6GEE45 pKa = 4.44YY46 pKa = 9.08KK47 pKa = 9.1TQIVKK52 pKa = 10.34QIIPQLCRR60 pKa = 11.84IPNQNSPPPIWVQGPRR76 pKa = 11.84IKK78 pKa = 10.85GDD80 pKa = 3.67PTWLKK85 pKa = 10.71INAKK89 pKa = 10.0FITEE93 pKa = 5.42LIPKK97 pKa = 9.79QKK99 pKa = 8.72GTKK102 pKa = 9.81NISTKK107 pKa = 10.15TYY109 pKa = 9.75LSRR112 pKa = 11.84LFVIWLQNN120 pKa = 3.18
Molecular weight: 14.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4217 |
120 |
1752 |
702.8 |
79.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.758 ± 0.709 | 1.66 ± 0.142 |
4.34 ± 0.461 | 5.193 ± 0.64 |
2.988 ± 0.25 | 4.743 ± 0.186 |
3.106 ± 0.316 | 7.209 ± 0.595 |
5.834 ± 0.921 | 9.153 ± 0.711 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.537 ± 0.292 | 4.387 ± 0.571 |
6.995 ± 0.38 | 6.782 ± 0.748 |
4.387 ± 0.219 | 5.762 ± 0.41 |
8.229 ± 0.446 | 5.478 ± 0.311 |
1.613 ± 0.137 | 2.822 ± 0.237 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
