
Hubei rhabdo-like virus 1
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.06
Get precalculated fractions of proteins
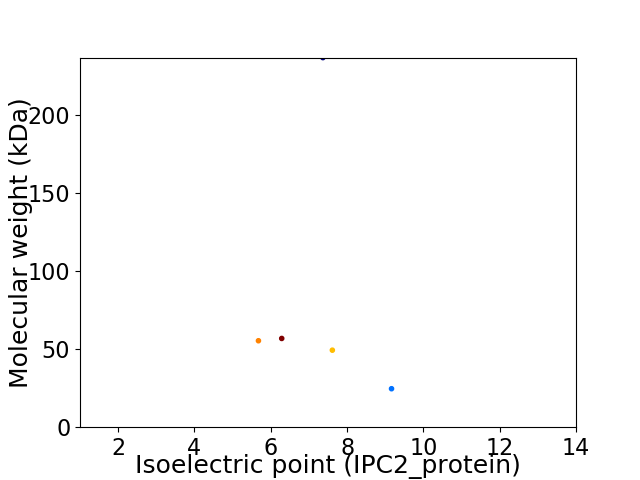
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KMW7|A0A1L3KMW7_9VIRU Uncharacterized protein OS=Hubei rhabdo-like virus 1 OX=1923185 PE=4 SV=1
MM1 pKa = 7.16YY2 pKa = 9.54PSEE5 pKa = 4.16TRR7 pKa = 11.84ILIATVIIIAIFGIIIILFCWCLAKK32 pKa = 10.12RR33 pKa = 11.84RR34 pKa = 11.84KK35 pKa = 9.24PGHH38 pKa = 6.34HH39 pKa = 6.35LTRR42 pKa = 11.84PHH44 pKa = 6.48RR45 pKa = 11.84CKK47 pKa = 10.22FHH49 pKa = 5.88YY50 pKa = 10.31SRR52 pKa = 11.84LRR54 pKa = 11.84DD55 pKa = 3.64LEE57 pKa = 4.11NQRR60 pKa = 11.84VTSGSSSAATSYY72 pKa = 11.24VSTPRR77 pKa = 11.84YY78 pKa = 8.55PADD81 pKa = 3.6SDD83 pKa = 3.78SSRR86 pKa = 11.84SVLTRR91 pKa = 11.84RR92 pKa = 11.84MSSLSDD98 pKa = 3.26KK99 pKa = 10.6LRR101 pKa = 11.84QIQEE105 pKa = 3.9EE106 pKa = 4.21DD107 pKa = 3.63PEE109 pKa = 4.37LKK111 pKa = 10.37RR112 pKa = 11.84RR113 pKa = 11.84FEE115 pKa = 4.38EE116 pKa = 3.89IKK118 pKa = 10.54IRR120 pKa = 11.84GADD123 pKa = 3.69SLSTPSDD130 pKa = 3.27ADD132 pKa = 3.81DD133 pKa = 3.7LVQRR137 pKa = 11.84SLRR140 pKa = 11.84GFSSEE145 pKa = 4.71SEE147 pKa = 3.63ILYY150 pKa = 10.37SLNYY154 pKa = 9.8LPSDD158 pKa = 3.9LEE160 pKa = 4.16VEE162 pKa = 4.53AGVEE166 pKa = 4.15TQRR169 pKa = 11.84TGEE172 pKa = 4.0GDD174 pKa = 3.26NTGGGTAGSSQSTSTQGDD192 pKa = 4.21CGPVLDD198 pKa = 5.37PYY200 pKa = 9.5WAAVLNSNFNQGHH213 pKa = 6.29PRR215 pKa = 11.84DD216 pKa = 3.6HH217 pKa = 6.4LQRR220 pKa = 11.84LGALAKK226 pKa = 10.24DD227 pKa = 4.33HH228 pKa = 7.67PDD230 pKa = 3.28PDD232 pKa = 3.75IPNIWSEE239 pKa = 3.93ITPIFQDD246 pKa = 3.78LLKK249 pKa = 10.84ANQIMLLRR257 pKa = 11.84LLLGEE262 pKa = 4.65LRR264 pKa = 11.84NPVQDD269 pKa = 2.05IRR271 pKa = 11.84MFIRR275 pKa = 11.84GVQFGHH281 pKa = 5.96QMRR284 pKa = 11.84EE285 pKa = 3.82YY286 pKa = 11.09SLLQAHH292 pKa = 5.94TLHH295 pKa = 6.74LEE297 pKa = 4.06SLIDD301 pKa = 3.54TMNVNNAKK309 pKa = 10.09VVGRR313 pKa = 11.84LEE315 pKa = 4.57ASMNQNAALASQVTEE330 pKa = 3.6ASKK333 pKa = 11.19NVTHH337 pKa = 6.86FVAAVEE343 pKa = 4.17KK344 pKa = 10.62AQLLAEE350 pKa = 4.23KK351 pKa = 9.68MEE353 pKa = 4.68LSLVPPEE360 pKa = 4.08VVPASKK366 pKa = 10.11HH367 pKa = 4.83AAPSIISRR375 pKa = 11.84VTEE378 pKa = 3.9TQKK381 pKa = 11.42AYY383 pKa = 9.37VTLDD387 pKa = 3.01AKK389 pKa = 11.03LGDD392 pKa = 3.7GVYY395 pKa = 9.94EE396 pKa = 4.25SKK398 pKa = 10.5IGLVTIVAEE407 pKa = 4.51KK408 pKa = 9.6VRR410 pKa = 11.84SVAIHH415 pKa = 6.87AGLNDD420 pKa = 3.24PWMPLVGMDD429 pKa = 3.5ARR431 pKa = 11.84VFQIMACVNLDD442 pKa = 3.57TLEE445 pKa = 4.94SKK447 pKa = 10.35LQPGVLASIFTGSSEE462 pKa = 4.11EE463 pKa = 4.16KK464 pKa = 9.93NSKK467 pKa = 10.09FGALIGSCGSAAMQWWFATRR487 pKa = 11.84VDD489 pKa = 4.0EE490 pKa = 5.56SMHH493 pKa = 7.15DD494 pKa = 3.91DD495 pKa = 3.51LSGEE499 pKa = 4.21SSSAA503 pKa = 3.16
MM1 pKa = 7.16YY2 pKa = 9.54PSEE5 pKa = 4.16TRR7 pKa = 11.84ILIATVIIIAIFGIIIILFCWCLAKK32 pKa = 10.12RR33 pKa = 11.84RR34 pKa = 11.84KK35 pKa = 9.24PGHH38 pKa = 6.34HH39 pKa = 6.35LTRR42 pKa = 11.84PHH44 pKa = 6.48RR45 pKa = 11.84CKK47 pKa = 10.22FHH49 pKa = 5.88YY50 pKa = 10.31SRR52 pKa = 11.84LRR54 pKa = 11.84DD55 pKa = 3.64LEE57 pKa = 4.11NQRR60 pKa = 11.84VTSGSSSAATSYY72 pKa = 11.24VSTPRR77 pKa = 11.84YY78 pKa = 8.55PADD81 pKa = 3.6SDD83 pKa = 3.78SSRR86 pKa = 11.84SVLTRR91 pKa = 11.84RR92 pKa = 11.84MSSLSDD98 pKa = 3.26KK99 pKa = 10.6LRR101 pKa = 11.84QIQEE105 pKa = 3.9EE106 pKa = 4.21DD107 pKa = 3.63PEE109 pKa = 4.37LKK111 pKa = 10.37RR112 pKa = 11.84RR113 pKa = 11.84FEE115 pKa = 4.38EE116 pKa = 3.89IKK118 pKa = 10.54IRR120 pKa = 11.84GADD123 pKa = 3.69SLSTPSDD130 pKa = 3.27ADD132 pKa = 3.81DD133 pKa = 3.7LVQRR137 pKa = 11.84SLRR140 pKa = 11.84GFSSEE145 pKa = 4.71SEE147 pKa = 3.63ILYY150 pKa = 10.37SLNYY154 pKa = 9.8LPSDD158 pKa = 3.9LEE160 pKa = 4.16VEE162 pKa = 4.53AGVEE166 pKa = 4.15TQRR169 pKa = 11.84TGEE172 pKa = 4.0GDD174 pKa = 3.26NTGGGTAGSSQSTSTQGDD192 pKa = 4.21CGPVLDD198 pKa = 5.37PYY200 pKa = 9.5WAAVLNSNFNQGHH213 pKa = 6.29PRR215 pKa = 11.84DD216 pKa = 3.6HH217 pKa = 6.4LQRR220 pKa = 11.84LGALAKK226 pKa = 10.24DD227 pKa = 4.33HH228 pKa = 7.67PDD230 pKa = 3.28PDD232 pKa = 3.75IPNIWSEE239 pKa = 3.93ITPIFQDD246 pKa = 3.78LLKK249 pKa = 10.84ANQIMLLRR257 pKa = 11.84LLLGEE262 pKa = 4.65LRR264 pKa = 11.84NPVQDD269 pKa = 2.05IRR271 pKa = 11.84MFIRR275 pKa = 11.84GVQFGHH281 pKa = 5.96QMRR284 pKa = 11.84EE285 pKa = 3.82YY286 pKa = 11.09SLLQAHH292 pKa = 5.94TLHH295 pKa = 6.74LEE297 pKa = 4.06SLIDD301 pKa = 3.54TMNVNNAKK309 pKa = 10.09VVGRR313 pKa = 11.84LEE315 pKa = 4.57ASMNQNAALASQVTEE330 pKa = 3.6ASKK333 pKa = 11.19NVTHH337 pKa = 6.86FVAAVEE343 pKa = 4.17KK344 pKa = 10.62AQLLAEE350 pKa = 4.23KK351 pKa = 9.68MEE353 pKa = 4.68LSLVPPEE360 pKa = 4.08VVPASKK366 pKa = 10.11HH367 pKa = 4.83AAPSIISRR375 pKa = 11.84VTEE378 pKa = 3.9TQKK381 pKa = 11.42AYY383 pKa = 9.37VTLDD387 pKa = 3.01AKK389 pKa = 11.03LGDD392 pKa = 3.7GVYY395 pKa = 9.94EE396 pKa = 4.25SKK398 pKa = 10.5IGLVTIVAEE407 pKa = 4.51KK408 pKa = 9.6VRR410 pKa = 11.84SVAIHH415 pKa = 6.87AGLNDD420 pKa = 3.24PWMPLVGMDD429 pKa = 3.5ARR431 pKa = 11.84VFQIMACVNLDD442 pKa = 3.57TLEE445 pKa = 4.94SKK447 pKa = 10.35LQPGVLASIFTGSSEE462 pKa = 4.11EE463 pKa = 4.16KK464 pKa = 9.93NSKK467 pKa = 10.09FGALIGSCGSAAMQWWFATRR487 pKa = 11.84VDD489 pKa = 4.0EE490 pKa = 5.56SMHH493 pKa = 7.15DD494 pKa = 3.91DD495 pKa = 3.51LSGEE499 pKa = 4.21SSSAA503 pKa = 3.16
Molecular weight: 55.36 kDa
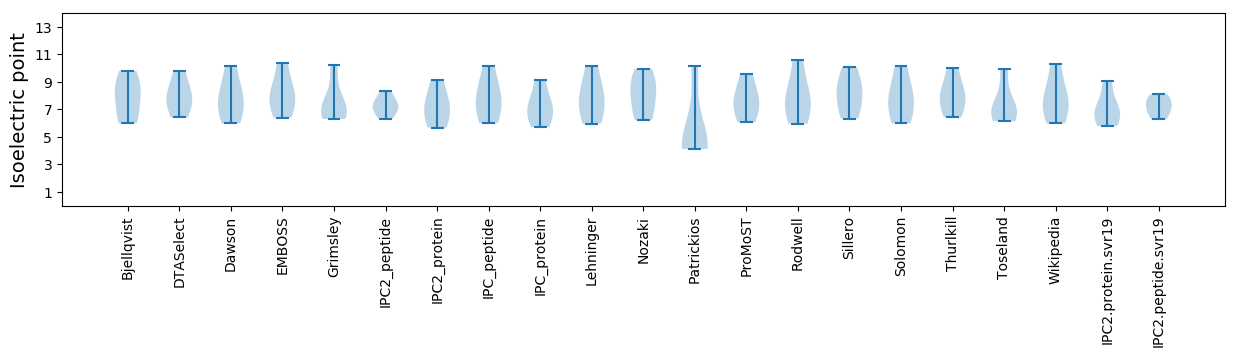
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KMV5|A0A1L3KMV5_9VIRU Uncharacterized protein OS=Hubei rhabdo-like virus 1 OX=1923185 PE=4 SV=1
MM1 pKa = 7.4TNLIRR6 pKa = 11.84RR7 pKa = 11.84FTRR10 pKa = 11.84SSKK13 pKa = 10.7ANKK16 pKa = 9.4VDD18 pKa = 3.58PQNNSPYY25 pKa = 9.32PPKK28 pKa = 10.53SFKK31 pKa = 9.83FTLEE35 pKa = 4.06HH36 pKa = 5.82IRR38 pKa = 11.84AAVALEE44 pKa = 3.75GVINFDD50 pKa = 3.0IEE52 pKa = 4.51GPIEE56 pKa = 4.13YY57 pKa = 9.25TVIPYY62 pKa = 9.91ISKK65 pKa = 10.66RR66 pKa = 11.84MVAYY70 pKa = 9.81ILRR73 pKa = 11.84IAIDD77 pKa = 3.04QHH79 pKa = 7.97IITEE83 pKa = 4.12EE84 pKa = 3.55AATYY88 pKa = 10.5FDD90 pKa = 3.66APLRR94 pKa = 11.84KK95 pKa = 9.61AIIQSAPYY103 pKa = 8.3QTSKK107 pKa = 10.63EE108 pKa = 3.94KK109 pKa = 10.88KK110 pKa = 8.53LIHH113 pKa = 7.1RR114 pKa = 11.84ITGVAPVKK122 pKa = 10.58LLTPVSCKK130 pKa = 9.9VVADD134 pKa = 4.18KK135 pKa = 11.3EE136 pKa = 4.48SLTWDD141 pKa = 3.56KK142 pKa = 8.39TTEE145 pKa = 4.18MIGKK149 pKa = 9.19ALYY152 pKa = 9.42PVVIRR157 pKa = 11.84CFGEE161 pKa = 4.36LNTSSFLLRR170 pKa = 11.84DD171 pKa = 3.37RR172 pKa = 11.84GLLRR176 pKa = 11.84GRR178 pKa = 11.84MFLSTPIPSPQQVAKK193 pKa = 9.32TKK195 pKa = 10.06SAIRR199 pKa = 11.84KK200 pKa = 7.17EE201 pKa = 4.03RR202 pKa = 11.84ARR204 pKa = 11.84ALAIEE209 pKa = 4.48DD210 pKa = 4.17TPGPSAPTLDD220 pKa = 3.44
MM1 pKa = 7.4TNLIRR6 pKa = 11.84RR7 pKa = 11.84FTRR10 pKa = 11.84SSKK13 pKa = 10.7ANKK16 pKa = 9.4VDD18 pKa = 3.58PQNNSPYY25 pKa = 9.32PPKK28 pKa = 10.53SFKK31 pKa = 9.83FTLEE35 pKa = 4.06HH36 pKa = 5.82IRR38 pKa = 11.84AAVALEE44 pKa = 3.75GVINFDD50 pKa = 3.0IEE52 pKa = 4.51GPIEE56 pKa = 4.13YY57 pKa = 9.25TVIPYY62 pKa = 9.91ISKK65 pKa = 10.66RR66 pKa = 11.84MVAYY70 pKa = 9.81ILRR73 pKa = 11.84IAIDD77 pKa = 3.04QHH79 pKa = 7.97IITEE83 pKa = 4.12EE84 pKa = 3.55AATYY88 pKa = 10.5FDD90 pKa = 3.66APLRR94 pKa = 11.84KK95 pKa = 9.61AIIQSAPYY103 pKa = 8.3QTSKK107 pKa = 10.63EE108 pKa = 3.94KK109 pKa = 10.88KK110 pKa = 8.53LIHH113 pKa = 7.1RR114 pKa = 11.84ITGVAPVKK122 pKa = 10.58LLTPVSCKK130 pKa = 9.9VVADD134 pKa = 4.18KK135 pKa = 11.3EE136 pKa = 4.48SLTWDD141 pKa = 3.56KK142 pKa = 8.39TTEE145 pKa = 4.18MIGKK149 pKa = 9.19ALYY152 pKa = 9.42PVVIRR157 pKa = 11.84CFGEE161 pKa = 4.36LNTSSFLLRR170 pKa = 11.84DD171 pKa = 3.37RR172 pKa = 11.84GLLRR176 pKa = 11.84GRR178 pKa = 11.84MFLSTPIPSPQQVAKK193 pKa = 9.32TKK195 pKa = 10.06SAIRR199 pKa = 11.84KK200 pKa = 7.17EE201 pKa = 4.03RR202 pKa = 11.84ARR204 pKa = 11.84ALAIEE209 pKa = 4.48DD210 pKa = 4.17TPGPSAPTLDD220 pKa = 3.44
Molecular weight: 24.64 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3790 |
220 |
2112 |
758.0 |
84.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.623 ± 0.863 | 1.319 ± 0.161 |
4.538 ± 0.227 | 5.858 ± 0.139 |
4.195 ± 0.345 | 6.332 ± 0.514 |
2.559 ± 0.283 | 6.174 ± 0.512 |
4.855 ± 0.413 | 10.712 ± 0.559 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.216 ± 0.165 | 3.43 ± 0.236 |
5.778 ± 0.346 | 3.562 ± 0.227 |
5.884 ± 0.275 | 9.024 ± 0.593 |
6.332 ± 0.338 | 6.332 ± 0.231 |
1.504 ± 0.155 | 2.77 ± 0.22 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
