
Mount Mabu Lophuromys virus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; Orthoparamyxovirinae; Jeilongvirus; Lophuromys jeilongvirus 1
Average proteome isoelectric point is 6.8
Get precalculated fractions of proteins
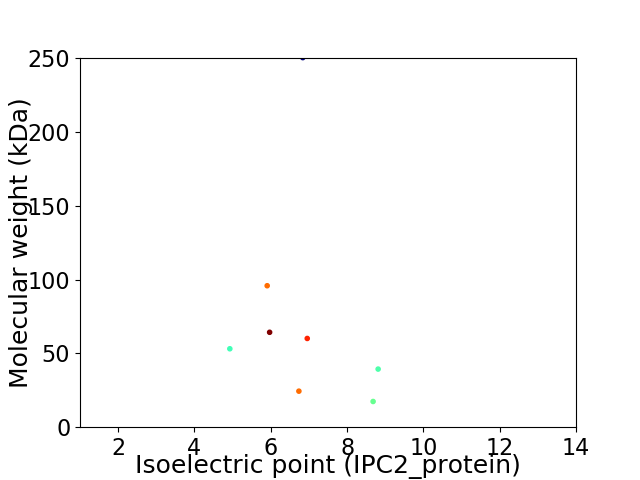
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A2P1GJ83|A0A2P1GJ83_9MONO Nucleocapsid OS=Mount Mabu Lophuromys virus 1 OX=2116559 GN=N PE=3 SV=1
MM1 pKa = 7.7AGVDD5 pKa = 3.95PNLDD9 pKa = 4.27LINNGIQTIEE19 pKa = 4.68FIQSNRR25 pKa = 11.84DD26 pKa = 3.71EE27 pKa = 4.17IQKK30 pKa = 9.55TYY32 pKa = 10.32GRR34 pKa = 11.84SSIGQPRR41 pKa = 11.84TKK43 pKa = 10.11EE44 pKa = 3.38RR45 pKa = 11.84AAAWEE50 pKa = 4.64AYY52 pKa = 10.29LEE54 pKa = 4.55SKK56 pKa = 10.51DD57 pKa = 3.76GHH59 pKa = 6.49PKK61 pKa = 8.24GQRR64 pKa = 11.84RR65 pKa = 11.84GGEE68 pKa = 4.19SQTSDD73 pKa = 3.44SEE75 pKa = 4.43SSSRR79 pKa = 11.84EE80 pKa = 3.91GNSDD84 pKa = 3.41LGSSLSQSSLSEE96 pKa = 3.73INPRR100 pKa = 11.84EE101 pKa = 4.09STGSGNYY108 pKa = 9.47KK109 pKa = 10.25GGYY112 pKa = 10.01DD113 pKa = 3.7EE114 pKa = 6.34DD115 pKa = 4.11PAEE118 pKa = 4.14LHH120 pKa = 6.53SGTRR124 pKa = 11.84DD125 pKa = 3.25DD126 pKa = 5.93CRR128 pKa = 11.84RR129 pKa = 11.84EE130 pKa = 4.02SNRR133 pKa = 11.84NGGGGTIIGEE143 pKa = 4.67GTDD146 pKa = 3.44STQGGNSSHH155 pKa = 6.89EE156 pKa = 4.25IDD158 pKa = 4.97DD159 pKa = 3.88GDD161 pKa = 3.8YY162 pKa = 11.55DD163 pKa = 5.21RR164 pKa = 11.84ICLLDD169 pKa = 3.64QATEE173 pKa = 3.72DD174 pKa = 3.56HH175 pKa = 6.62LAGKK179 pKa = 7.43EE180 pKa = 4.26TPGKK184 pKa = 10.55LEE186 pKa = 3.69VRR188 pKa = 11.84EE189 pKa = 4.42ARR191 pKa = 11.84PSDD194 pKa = 3.04IAEE197 pKa = 4.26ILSEE201 pKa = 4.32DD202 pKa = 3.16SSRR205 pKa = 11.84THH207 pKa = 5.86RR208 pKa = 11.84RR209 pKa = 11.84LRR211 pKa = 11.84GIRR214 pKa = 11.84SVSRR218 pKa = 11.84EE219 pKa = 4.16SEE221 pKa = 4.22SEE223 pKa = 4.0EE224 pKa = 4.45SDD226 pKa = 3.39VSPVKK231 pKa = 10.55KK232 pKa = 10.46GIAGSIVSMSSKK244 pKa = 10.47GKK246 pKa = 7.92HH247 pKa = 4.78TSGSGATQHH256 pKa = 6.78ALLSPSTPTNRR267 pKa = 11.84SVSADD272 pKa = 3.54DD273 pKa = 3.79ARR275 pKa = 11.84EE276 pKa = 3.91SARR279 pKa = 11.84TARR282 pKa = 11.84EE283 pKa = 3.26ISIDD287 pKa = 3.54DD288 pKa = 3.85DD289 pKa = 4.46LGVFSSFGGGSDD301 pKa = 3.18ISDD304 pKa = 4.03KK305 pKa = 10.37IDD307 pKa = 4.13RR308 pKa = 11.84MLEE311 pKa = 3.91NQNEE315 pKa = 4.41ILSKK319 pKa = 10.44LQVLAEE325 pKa = 3.93VSEE328 pKa = 4.43EE329 pKa = 3.56IKK331 pKa = 10.81GIKK334 pKa = 9.62KK335 pKa = 9.99ILTNHH340 pKa = 6.04SLSLSTLEE348 pKa = 5.39GYY350 pKa = 10.35INDD353 pKa = 3.97LMIVIPKK360 pKa = 10.22SGISNSDD367 pKa = 2.95GDD369 pKa = 4.44DD370 pKa = 3.43EE371 pKa = 5.19KK372 pKa = 11.67NPDD375 pKa = 3.14LRR377 pKa = 11.84MVVGRR382 pKa = 11.84DD383 pKa = 2.97KK384 pKa = 11.4SRR386 pKa = 11.84GVTEE390 pKa = 3.59VSKK393 pKa = 10.82RR394 pKa = 11.84KK395 pKa = 9.54RR396 pKa = 11.84PKK398 pKa = 8.69STFGEE403 pKa = 4.41EE404 pKa = 3.47IGTGRR409 pKa = 11.84TIDD412 pKa = 3.7EE413 pKa = 4.89EE414 pKa = 4.51YY415 pKa = 11.26LLDD418 pKa = 4.64PLDD421 pKa = 4.02FKK423 pKa = 11.6KK424 pKa = 10.81NNAANFVPDD433 pKa = 3.71NTPVSMSIIASMVKK447 pKa = 10.11KK448 pKa = 10.16RR449 pKa = 11.84VHH451 pKa = 7.11DD452 pKa = 4.27PDD454 pKa = 3.74TRR456 pKa = 11.84DD457 pKa = 3.34EE458 pKa = 4.52LLMLVEE464 pKa = 5.02SNLGSIPMKK473 pKa = 10.77DD474 pKa = 3.02MYY476 pKa = 10.85EE477 pKa = 4.13QIRR480 pKa = 11.84KK481 pKa = 9.5HH482 pKa = 5.96IDD484 pKa = 3.13QMDD487 pKa = 3.37SS488 pKa = 2.94
MM1 pKa = 7.7AGVDD5 pKa = 3.95PNLDD9 pKa = 4.27LINNGIQTIEE19 pKa = 4.68FIQSNRR25 pKa = 11.84DD26 pKa = 3.71EE27 pKa = 4.17IQKK30 pKa = 9.55TYY32 pKa = 10.32GRR34 pKa = 11.84SSIGQPRR41 pKa = 11.84TKK43 pKa = 10.11EE44 pKa = 3.38RR45 pKa = 11.84AAAWEE50 pKa = 4.64AYY52 pKa = 10.29LEE54 pKa = 4.55SKK56 pKa = 10.51DD57 pKa = 3.76GHH59 pKa = 6.49PKK61 pKa = 8.24GQRR64 pKa = 11.84RR65 pKa = 11.84GGEE68 pKa = 4.19SQTSDD73 pKa = 3.44SEE75 pKa = 4.43SSSRR79 pKa = 11.84EE80 pKa = 3.91GNSDD84 pKa = 3.41LGSSLSQSSLSEE96 pKa = 3.73INPRR100 pKa = 11.84EE101 pKa = 4.09STGSGNYY108 pKa = 9.47KK109 pKa = 10.25GGYY112 pKa = 10.01DD113 pKa = 3.7EE114 pKa = 6.34DD115 pKa = 4.11PAEE118 pKa = 4.14LHH120 pKa = 6.53SGTRR124 pKa = 11.84DD125 pKa = 3.25DD126 pKa = 5.93CRR128 pKa = 11.84RR129 pKa = 11.84EE130 pKa = 4.02SNRR133 pKa = 11.84NGGGGTIIGEE143 pKa = 4.67GTDD146 pKa = 3.44STQGGNSSHH155 pKa = 6.89EE156 pKa = 4.25IDD158 pKa = 4.97DD159 pKa = 3.88GDD161 pKa = 3.8YY162 pKa = 11.55DD163 pKa = 5.21RR164 pKa = 11.84ICLLDD169 pKa = 3.64QATEE173 pKa = 3.72DD174 pKa = 3.56HH175 pKa = 6.62LAGKK179 pKa = 7.43EE180 pKa = 4.26TPGKK184 pKa = 10.55LEE186 pKa = 3.69VRR188 pKa = 11.84EE189 pKa = 4.42ARR191 pKa = 11.84PSDD194 pKa = 3.04IAEE197 pKa = 4.26ILSEE201 pKa = 4.32DD202 pKa = 3.16SSRR205 pKa = 11.84THH207 pKa = 5.86RR208 pKa = 11.84RR209 pKa = 11.84LRR211 pKa = 11.84GIRR214 pKa = 11.84SVSRR218 pKa = 11.84EE219 pKa = 4.16SEE221 pKa = 4.22SEE223 pKa = 4.0EE224 pKa = 4.45SDD226 pKa = 3.39VSPVKK231 pKa = 10.55KK232 pKa = 10.46GIAGSIVSMSSKK244 pKa = 10.47GKK246 pKa = 7.92HH247 pKa = 4.78TSGSGATQHH256 pKa = 6.78ALLSPSTPTNRR267 pKa = 11.84SVSADD272 pKa = 3.54DD273 pKa = 3.79ARR275 pKa = 11.84EE276 pKa = 3.91SARR279 pKa = 11.84TARR282 pKa = 11.84EE283 pKa = 3.26ISIDD287 pKa = 3.54DD288 pKa = 3.85DD289 pKa = 4.46LGVFSSFGGGSDD301 pKa = 3.18ISDD304 pKa = 4.03KK305 pKa = 10.37IDD307 pKa = 4.13RR308 pKa = 11.84MLEE311 pKa = 3.91NQNEE315 pKa = 4.41ILSKK319 pKa = 10.44LQVLAEE325 pKa = 3.93VSEE328 pKa = 4.43EE329 pKa = 3.56IKK331 pKa = 10.81GIKK334 pKa = 9.62KK335 pKa = 9.99ILTNHH340 pKa = 6.04SLSLSTLEE348 pKa = 5.39GYY350 pKa = 10.35INDD353 pKa = 3.97LMIVIPKK360 pKa = 10.22SGISNSDD367 pKa = 2.95GDD369 pKa = 4.44DD370 pKa = 3.43EE371 pKa = 5.19KK372 pKa = 11.67NPDD375 pKa = 3.14LRR377 pKa = 11.84MVVGRR382 pKa = 11.84DD383 pKa = 2.97KK384 pKa = 11.4SRR386 pKa = 11.84GVTEE390 pKa = 3.59VSKK393 pKa = 10.82RR394 pKa = 11.84KK395 pKa = 9.54RR396 pKa = 11.84PKK398 pKa = 8.69STFGEE403 pKa = 4.41EE404 pKa = 3.47IGTGRR409 pKa = 11.84TIDD412 pKa = 3.7EE413 pKa = 4.89EE414 pKa = 4.51YY415 pKa = 11.26LLDD418 pKa = 4.64PLDD421 pKa = 4.02FKK423 pKa = 11.6KK424 pKa = 10.81NNAANFVPDD433 pKa = 3.71NTPVSMSIIASMVKK447 pKa = 10.11KK448 pKa = 10.16RR449 pKa = 11.84VHH451 pKa = 7.11DD452 pKa = 4.27PDD454 pKa = 3.74TRR456 pKa = 11.84DD457 pKa = 3.34EE458 pKa = 4.52LLMLVEE464 pKa = 5.02SNLGSIPMKK473 pKa = 10.77DD474 pKa = 3.02MYY476 pKa = 10.85EE477 pKa = 4.13QIRR480 pKa = 11.84KK481 pKa = 9.5HH482 pKa = 5.96IDD484 pKa = 3.13QMDD487 pKa = 3.37SS488 pKa = 2.94
Molecular weight: 53.16 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2P1GJA5|A0A2P1GJA5_9MONO Matrix protein OS=Mount Mabu Lophuromys virus 1 OX=2116559 GN=M PE=4 SV=1
MM1 pKa = 7.46YY2 pKa = 10.18YY3 pKa = 10.28KK4 pKa = 10.69KK5 pKa = 10.7LRR7 pKa = 11.84IKK9 pKa = 10.18VIHH12 pKa = 6.11ITMEE16 pKa = 4.1GKK18 pKa = 10.45ADD20 pKa = 4.19FLPSSWSEE28 pKa = 3.59GGILHH33 pKa = 7.39AIEE36 pKa = 5.77AEE38 pKa = 3.95ADD40 pKa = 3.79DD41 pKa = 4.46EE42 pKa = 4.8GKK44 pKa = 10.45LVPKK48 pKa = 10.29YY49 pKa = 10.37RR50 pKa = 11.84VVNPGWNSRR59 pKa = 11.84KK60 pKa = 10.14GSGFMYY66 pKa = 10.6LLIYY70 pKa = 9.71GIIEE74 pKa = 4.06EE75 pKa = 4.95KK76 pKa = 10.34EE77 pKa = 4.24SKK79 pKa = 10.24DD80 pKa = 3.21KK81 pKa = 11.12KK82 pKa = 10.54NRR84 pKa = 11.84GFKK87 pKa = 10.73SFGAFPLGVGRR98 pKa = 11.84SRR100 pKa = 11.84ADD102 pKa = 3.38PQDD105 pKa = 3.92LLDD108 pKa = 5.57AVLQLDD114 pKa = 3.34ITVRR118 pKa = 11.84RR119 pKa = 11.84TAGAGEE125 pKa = 4.27KK126 pKa = 10.54LVMGTNNINPILTPWKK142 pKa = 10.52GILTNGAVFQAIKK155 pKa = 10.33VCNNVDD161 pKa = 3.72TIMVDD166 pKa = 3.4RR167 pKa = 11.84PQKK170 pKa = 10.24LRR172 pKa = 11.84PIFLTITLLTDD183 pKa = 3.93AGCYY187 pKa = 9.55KK188 pKa = 10.35IPRR191 pKa = 11.84TILDD195 pKa = 3.74FRR197 pKa = 11.84ASGAMSFNLLVVLSIGADD215 pKa = 3.44FTPYY219 pKa = 8.34GTKK222 pKa = 10.78GMINEE227 pKa = 4.52DD228 pKa = 3.87GEE230 pKa = 4.55RR231 pKa = 11.84VTTFMVHH238 pKa = 5.85IGNFVRR244 pKa = 11.84RR245 pKa = 11.84CGKK248 pKa = 9.89EE249 pKa = 3.61YY250 pKa = 10.9SVDD253 pKa = 3.38YY254 pKa = 10.77CRR256 pKa = 11.84QKK258 pKa = 10.21VDD260 pKa = 3.46KK261 pKa = 9.78MDD263 pKa = 3.58LRR265 pKa = 11.84FSLGAVGGLSFHH277 pKa = 6.77VKK279 pKa = 10.21LAGKK283 pKa = 9.68VSKK286 pKa = 8.5TLRR289 pKa = 11.84AQLGHH294 pKa = 6.65RR295 pKa = 11.84TTICYY300 pKa = 10.8SLMDD304 pKa = 4.0TNPYY308 pKa = 9.65LNRR311 pKa = 11.84VMWKK315 pKa = 10.16AEE317 pKa = 3.75CSIQKK322 pKa = 8.05VTAVFQPSVPRR333 pKa = 11.84DD334 pKa = 3.23FKK336 pKa = 11.1IYY338 pKa = 10.6DD339 pKa = 3.84DD340 pKa = 4.07VLIDD344 pKa = 3.55HH345 pKa = 7.21TGKK348 pKa = 10.1ILKK351 pKa = 9.27QQ352 pKa = 3.26
MM1 pKa = 7.46YY2 pKa = 10.18YY3 pKa = 10.28KK4 pKa = 10.69KK5 pKa = 10.7LRR7 pKa = 11.84IKK9 pKa = 10.18VIHH12 pKa = 6.11ITMEE16 pKa = 4.1GKK18 pKa = 10.45ADD20 pKa = 4.19FLPSSWSEE28 pKa = 3.59GGILHH33 pKa = 7.39AIEE36 pKa = 5.77AEE38 pKa = 3.95ADD40 pKa = 3.79DD41 pKa = 4.46EE42 pKa = 4.8GKK44 pKa = 10.45LVPKK48 pKa = 10.29YY49 pKa = 10.37RR50 pKa = 11.84VVNPGWNSRR59 pKa = 11.84KK60 pKa = 10.14GSGFMYY66 pKa = 10.6LLIYY70 pKa = 9.71GIIEE74 pKa = 4.06EE75 pKa = 4.95KK76 pKa = 10.34EE77 pKa = 4.24SKK79 pKa = 10.24DD80 pKa = 3.21KK81 pKa = 11.12KK82 pKa = 10.54NRR84 pKa = 11.84GFKK87 pKa = 10.73SFGAFPLGVGRR98 pKa = 11.84SRR100 pKa = 11.84ADD102 pKa = 3.38PQDD105 pKa = 3.92LLDD108 pKa = 5.57AVLQLDD114 pKa = 3.34ITVRR118 pKa = 11.84RR119 pKa = 11.84TAGAGEE125 pKa = 4.27KK126 pKa = 10.54LVMGTNNINPILTPWKK142 pKa = 10.52GILTNGAVFQAIKK155 pKa = 10.33VCNNVDD161 pKa = 3.72TIMVDD166 pKa = 3.4RR167 pKa = 11.84PQKK170 pKa = 10.24LRR172 pKa = 11.84PIFLTITLLTDD183 pKa = 3.93AGCYY187 pKa = 9.55KK188 pKa = 10.35IPRR191 pKa = 11.84TILDD195 pKa = 3.74FRR197 pKa = 11.84ASGAMSFNLLVVLSIGADD215 pKa = 3.44FTPYY219 pKa = 8.34GTKK222 pKa = 10.78GMINEE227 pKa = 4.52DD228 pKa = 3.87GEE230 pKa = 4.55RR231 pKa = 11.84VTTFMVHH238 pKa = 5.85IGNFVRR244 pKa = 11.84RR245 pKa = 11.84CGKK248 pKa = 9.89EE249 pKa = 3.61YY250 pKa = 10.9SVDD253 pKa = 3.38YY254 pKa = 10.77CRR256 pKa = 11.84QKK258 pKa = 10.21VDD260 pKa = 3.46KK261 pKa = 9.78MDD263 pKa = 3.58LRR265 pKa = 11.84FSLGAVGGLSFHH277 pKa = 6.77VKK279 pKa = 10.21LAGKK283 pKa = 9.68VSKK286 pKa = 8.5TLRR289 pKa = 11.84AQLGHH294 pKa = 6.65RR295 pKa = 11.84TTICYY300 pKa = 10.8SLMDD304 pKa = 4.0TNPYY308 pKa = 9.65LNRR311 pKa = 11.84VMWKK315 pKa = 10.16AEE317 pKa = 3.75CSIQKK322 pKa = 8.05VTAVFQPSVPRR333 pKa = 11.84DD334 pKa = 3.23FKK336 pKa = 11.1IYY338 pKa = 10.6DD339 pKa = 3.84DD340 pKa = 4.07VLIDD344 pKa = 3.55HH345 pKa = 7.21TGKK348 pKa = 10.1ILKK351 pKa = 9.27QQ352 pKa = 3.26
Molecular weight: 39.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5363 |
155 |
2174 |
670.4 |
75.66 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.96 ± 0.639 | 1.417 ± 0.273 |
6.023 ± 0.36 | 5.519 ± 0.714 |
3.636 ± 0.512 | 5.948 ± 0.806 |
1.846 ± 0.396 | 7.869 ± 0.451 |
6.172 ± 0.373 | 9.528 ± 0.745 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.666 ± 0.238 | 5.762 ± 0.545 |
4.55 ± 0.439 | 3.841 ± 0.233 |
5.221 ± 0.51 | 8.186 ± 0.73 |
6.228 ± 0.771 | 5.668 ± 0.36 |
0.951 ± 0.178 | 4.009 ± 0.671 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
