
Salehabad virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Phenuiviridae; Phlebovirus; Salehabad phlebovirus
Average proteome isoelectric point is 6.51
Get precalculated fractions of proteins
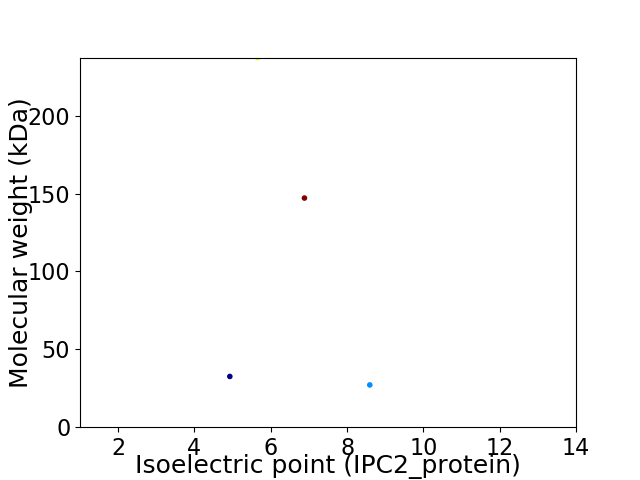
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|L0GAZ6|L0GAZ6_9VIRU Polyprotein OS=Salehabad virus OX=904699 PE=4 SV=1
TT1 pKa = 6.23KK2 pKa = 10.03TPYY5 pKa = 10.63CFIQTITMSLHH16 pKa = 5.06YY17 pKa = 10.21LCDD20 pKa = 5.38RR21 pKa = 11.84ITPTYY26 pKa = 10.39DD27 pKa = 3.39YY28 pKa = 9.79PCLGEE33 pKa = 4.06VMVSFEE39 pKa = 4.35AVNKK43 pKa = 9.65EE44 pKa = 3.74VGGPVCVYY52 pKa = 11.17DD53 pKa = 4.3DD54 pKa = 4.99LEE56 pKa = 5.4FEE58 pKa = 4.66LFEE61 pKa = 4.48SRR63 pKa = 11.84QSLSYY68 pKa = 11.05RR69 pKa = 11.84NTLYY73 pKa = 11.08DD74 pKa = 4.7FIDD77 pKa = 3.8SGEE80 pKa = 4.18LPWRR84 pKa = 11.84WGPHH88 pKa = 3.66MWGSRR93 pKa = 11.84VTLDD97 pKa = 4.05EE98 pKa = 5.29LPLLDD103 pKa = 5.04SLMKK107 pKa = 9.94TFQGLPVSDD116 pKa = 4.55LMDD119 pKa = 3.65SRR121 pKa = 11.84LHH123 pKa = 4.44YY124 pKa = 9.53TRR126 pKa = 11.84NSLIWPTGHH135 pKa = 6.49VSLKK139 pKa = 10.35AFRR142 pKa = 11.84ILYY145 pKa = 8.55PCIFVSFSYY154 pKa = 10.43SRR156 pKa = 11.84RR157 pKa = 11.84EE158 pKa = 3.38LAEE161 pKa = 4.09YY162 pKa = 10.15IFEE165 pKa = 4.34VTKK168 pKa = 10.75APHH171 pKa = 6.08LEE173 pKa = 3.93KK174 pKa = 10.83GIVDD178 pKa = 3.27MAGRR182 pKa = 11.84IRR184 pKa = 11.84SMAHH188 pKa = 5.26SLGIQEE194 pKa = 4.97NLVPCKK200 pKa = 11.11DD201 pKa = 3.54MMKK204 pKa = 10.16SICILQFFRR213 pKa = 11.84MIRR216 pKa = 11.84GFQRR220 pKa = 11.84DD221 pKa = 3.39RR222 pKa = 11.84TRR224 pKa = 11.84SVLFGGTVIPFFQACIDD241 pKa = 3.43ALTRR245 pKa = 11.84DD246 pKa = 4.17FPGLLDD252 pKa = 3.5VWSPGADD259 pKa = 2.59IFDD262 pKa = 5.98AITPSPCDD270 pKa = 3.22SGVSTDD276 pKa = 4.45SEE278 pKa = 4.22FDD280 pKa = 3.16SDD282 pKa = 4.69IEE284 pKa = 4.3VV285 pKa = 3.14
TT1 pKa = 6.23KK2 pKa = 10.03TPYY5 pKa = 10.63CFIQTITMSLHH16 pKa = 5.06YY17 pKa = 10.21LCDD20 pKa = 5.38RR21 pKa = 11.84ITPTYY26 pKa = 10.39DD27 pKa = 3.39YY28 pKa = 9.79PCLGEE33 pKa = 4.06VMVSFEE39 pKa = 4.35AVNKK43 pKa = 9.65EE44 pKa = 3.74VGGPVCVYY52 pKa = 11.17DD53 pKa = 4.3DD54 pKa = 4.99LEE56 pKa = 5.4FEE58 pKa = 4.66LFEE61 pKa = 4.48SRR63 pKa = 11.84QSLSYY68 pKa = 11.05RR69 pKa = 11.84NTLYY73 pKa = 11.08DD74 pKa = 4.7FIDD77 pKa = 3.8SGEE80 pKa = 4.18LPWRR84 pKa = 11.84WGPHH88 pKa = 3.66MWGSRR93 pKa = 11.84VTLDD97 pKa = 4.05EE98 pKa = 5.29LPLLDD103 pKa = 5.04SLMKK107 pKa = 9.94TFQGLPVSDD116 pKa = 4.55LMDD119 pKa = 3.65SRR121 pKa = 11.84LHH123 pKa = 4.44YY124 pKa = 9.53TRR126 pKa = 11.84NSLIWPTGHH135 pKa = 6.49VSLKK139 pKa = 10.35AFRR142 pKa = 11.84ILYY145 pKa = 8.55PCIFVSFSYY154 pKa = 10.43SRR156 pKa = 11.84RR157 pKa = 11.84EE158 pKa = 3.38LAEE161 pKa = 4.09YY162 pKa = 10.15IFEE165 pKa = 4.34VTKK168 pKa = 10.75APHH171 pKa = 6.08LEE173 pKa = 3.93KK174 pKa = 10.83GIVDD178 pKa = 3.27MAGRR182 pKa = 11.84IRR184 pKa = 11.84SMAHH188 pKa = 5.26SLGIQEE194 pKa = 4.97NLVPCKK200 pKa = 11.11DD201 pKa = 3.54MMKK204 pKa = 10.16SICILQFFRR213 pKa = 11.84MIRR216 pKa = 11.84GFQRR220 pKa = 11.84DD221 pKa = 3.39RR222 pKa = 11.84TRR224 pKa = 11.84SVLFGGTVIPFFQACIDD241 pKa = 3.43ALTRR245 pKa = 11.84DD246 pKa = 4.17FPGLLDD252 pKa = 3.5VWSPGADD259 pKa = 2.59IFDD262 pKa = 5.98AITPSPCDD270 pKa = 3.22SGVSTDD276 pKa = 4.45SEE278 pKa = 4.22FDD280 pKa = 3.16SDD282 pKa = 4.69IEE284 pKa = 4.3VV285 pKa = 3.14
Molecular weight: 32.59 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|L0GCI2|L0GCI2_9VIRU Replicase OS=Salehabad virus OX=904699 PE=4 SV=1
MM1 pKa = 7.57SGQEE5 pKa = 4.1EE6 pKa = 4.18FAKK9 pKa = 10.48IAVEE13 pKa = 4.25FANEE17 pKa = 4.02AVDD20 pKa = 3.51TAAILEE26 pKa = 4.32IVKK29 pKa = 9.88EE30 pKa = 4.17FAYY33 pKa = 10.15QGYY36 pKa = 9.05DD37 pKa = 2.88AARR40 pKa = 11.84VVEE43 pKa = 4.13LVRR46 pKa = 11.84EE47 pKa = 4.23KK48 pKa = 11.35GGDD51 pKa = 2.92TWRR54 pKa = 11.84NDD56 pKa = 2.98VKK58 pKa = 10.33TLIIIALTRR67 pKa = 11.84GNKK70 pKa = 6.23PTKK73 pKa = 9.71ILEE76 pKa = 4.37KK77 pKa = 9.66MSPAGKK83 pKa = 9.93SKK85 pKa = 10.12FRR87 pKa = 11.84ALVLKK92 pKa = 10.8YY93 pKa = 10.04SLKK96 pKa = 10.59SGNPGRR102 pKa = 11.84DD103 pKa = 3.46DD104 pKa = 3.39LTLARR109 pKa = 11.84IASAFAAWTIQAIKK123 pKa = 10.56VVEE126 pKa = 4.26QYY128 pKa = 11.5LPVTGAAMDD137 pKa = 4.4EE138 pKa = 4.55LSSSYY143 pKa = 9.23PRR145 pKa = 11.84PMMHH149 pKa = 7.01PCFGGLIDD157 pKa = 3.73NTLPEE162 pKa = 4.31EE163 pKa = 4.17TVAALVRR170 pKa = 11.84AHH172 pKa = 6.95SLFLDD177 pKa = 3.88TFSRR181 pKa = 11.84TINPAMRR188 pKa = 11.84TKK190 pKa = 10.42TKK192 pKa = 11.12AEE194 pKa = 3.89VLKK197 pKa = 11.18SFEE200 pKa = 4.21QPLNAAINSRR210 pKa = 11.84FLTSEE215 pKa = 4.4AKK217 pKa = 10.25RR218 pKa = 11.84KK219 pKa = 8.68ILKK222 pKa = 9.76SAGVIDD228 pKa = 4.51TNLKK232 pKa = 9.76PSPAVKK238 pKa = 9.99QAADD242 pKa = 3.77KK243 pKa = 10.85FSEE246 pKa = 4.5TAA248 pKa = 3.36
MM1 pKa = 7.57SGQEE5 pKa = 4.1EE6 pKa = 4.18FAKK9 pKa = 10.48IAVEE13 pKa = 4.25FANEE17 pKa = 4.02AVDD20 pKa = 3.51TAAILEE26 pKa = 4.32IVKK29 pKa = 9.88EE30 pKa = 4.17FAYY33 pKa = 10.15QGYY36 pKa = 9.05DD37 pKa = 2.88AARR40 pKa = 11.84VVEE43 pKa = 4.13LVRR46 pKa = 11.84EE47 pKa = 4.23KK48 pKa = 11.35GGDD51 pKa = 2.92TWRR54 pKa = 11.84NDD56 pKa = 2.98VKK58 pKa = 10.33TLIIIALTRR67 pKa = 11.84GNKK70 pKa = 6.23PTKK73 pKa = 9.71ILEE76 pKa = 4.37KK77 pKa = 9.66MSPAGKK83 pKa = 9.93SKK85 pKa = 10.12FRR87 pKa = 11.84ALVLKK92 pKa = 10.8YY93 pKa = 10.04SLKK96 pKa = 10.59SGNPGRR102 pKa = 11.84DD103 pKa = 3.46DD104 pKa = 3.39LTLARR109 pKa = 11.84IASAFAAWTIQAIKK123 pKa = 10.56VVEE126 pKa = 4.26QYY128 pKa = 11.5LPVTGAAMDD137 pKa = 4.4EE138 pKa = 4.55LSSSYY143 pKa = 9.23PRR145 pKa = 11.84PMMHH149 pKa = 7.01PCFGGLIDD157 pKa = 3.73NTLPEE162 pKa = 4.31EE163 pKa = 4.17TVAALVRR170 pKa = 11.84AHH172 pKa = 6.95SLFLDD177 pKa = 3.88TFSRR181 pKa = 11.84TINPAMRR188 pKa = 11.84TKK190 pKa = 10.42TKK192 pKa = 11.12AEE194 pKa = 3.89VLKK197 pKa = 11.18SFEE200 pKa = 4.21QPLNAAINSRR210 pKa = 11.84FLTSEE215 pKa = 4.4AKK217 pKa = 10.25RR218 pKa = 11.84KK219 pKa = 8.68ILKK222 pKa = 9.76SAGVIDD228 pKa = 4.51TNLKK232 pKa = 9.76PSPAVKK238 pKa = 9.99QAADD242 pKa = 3.77KK243 pKa = 10.85FSEE246 pKa = 4.5TAA248 pKa = 3.36
Molecular weight: 27.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3966 |
248 |
2096 |
991.5 |
111.07 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.606 ± 1.039 | 2.597 ± 0.957 |
5.497 ± 0.507 | 6.531 ± 0.609 |
4.438 ± 0.58 | 5.925 ± 0.415 |
2.32 ± 0.313 | 6.278 ± 0.116 |
5.799 ± 0.54 | 9.228 ± 0.12 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.454 ± 0.472 | 3.454 ± 0.399 |
4.387 ± 0.358 | 2.799 ± 0.118 |
5.371 ± 0.272 | 9.002 ± 0.527 |
5.799 ± 0.436 | 6.354 ± 0.295 |
1.286 ± 0.093 | 2.874 ± 0.174 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
