
Erinaceus europaeus papillomavirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Dyoetapapillomavirus; Dyoetapapillomavirus 1
Average proteome isoelectric point is 6.32
Get precalculated fractions of proteins
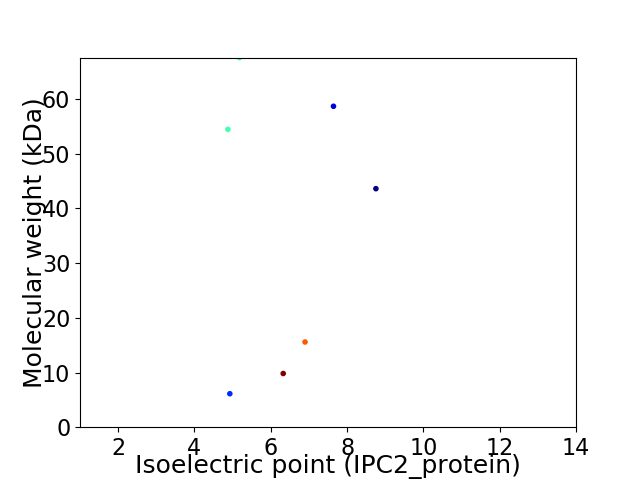
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B7TQP2|B7TQP2_9PAPI Minor capsid protein L2 OS=Erinaceus europaeus papillomavirus 1 OX=445217 GN=L2 PE=3 SV=1
MM1 pKa = 7.26ARR3 pKa = 11.84SRR5 pKa = 11.84RR6 pKa = 11.84TKK8 pKa = 10.16RR9 pKa = 11.84ASAQDD14 pKa = 3.87LYY16 pKa = 11.2KK17 pKa = 10.69SCLSGGDD24 pKa = 3.86CPPDD28 pKa = 3.34VKK30 pKa = 10.92NKK32 pKa = 10.19YY33 pKa = 7.32EE34 pKa = 4.07QNTTADD40 pKa = 4.17KK41 pKa = 10.18ILKK44 pKa = 9.13YY45 pKa = 10.75GSTAVYY51 pKa = 10.09FGGLGIGTGKK61 pKa = 8.82GTGGVTGYY69 pKa = 8.86TPLGGGEE76 pKa = 4.15GPVRR80 pKa = 11.84VGVPRR85 pKa = 11.84VLRR88 pKa = 11.84PALATDD94 pKa = 4.08LVGPLDD100 pKa = 4.04IAPVDD105 pKa = 3.79AGVGANDD112 pKa = 4.01PSVITLTDD120 pKa = 3.11STLSVDD126 pKa = 3.6VGPGEE131 pKa = 4.26VEE133 pKa = 4.4VVAEE137 pKa = 3.99IHH139 pKa = 6.19PVPDD143 pKa = 3.32TSTTTTGSGQGTSAVLEE160 pKa = 4.22ITPEE164 pKa = 3.79ATPGKK169 pKa = 9.74VRR171 pKa = 11.84VSRR174 pKa = 11.84TQYY177 pKa = 10.34HH178 pKa = 5.77NPAFHH183 pKa = 7.44ILTTSTPIGGEE194 pKa = 3.8TSAVDD199 pKa = 3.42NVFVDD204 pKa = 4.89FGSPSGHH211 pKa = 5.98IVGADD216 pKa = 3.15GVEE219 pKa = 4.34VYY221 pKa = 10.82EE222 pKa = 4.77EE223 pKa = 4.42IPLEE227 pKa = 3.92PLNRR231 pKa = 11.84SEE233 pKa = 5.98FEE235 pKa = 4.32IEE237 pKa = 3.91EE238 pKa = 4.05QTPKK242 pKa = 10.87SSTPKK247 pKa = 10.35ILDD250 pKa = 3.97TIADD254 pKa = 4.16SVRR257 pKa = 11.84KK258 pKa = 9.53FYY260 pKa = 11.07NRR262 pKa = 11.84RR263 pKa = 11.84IQQIKK268 pKa = 7.94TSDD271 pKa = 3.42YY272 pKa = 10.93RR273 pKa = 11.84FLSSPGKK280 pKa = 10.39LVDD283 pKa = 4.01ASFINPAYY291 pKa = 10.59DD292 pKa = 3.68PDD294 pKa = 3.71EE295 pKa = 4.23TLVFDD300 pKa = 3.88TDD302 pKa = 3.43INEE305 pKa = 4.17VQMAPNPDD313 pKa = 3.45FQDD316 pKa = 2.64IRR318 pKa = 11.84VLNRR322 pKa = 11.84PMFSTKK328 pKa = 9.67EE329 pKa = 3.8GYY331 pKa = 9.81IRR333 pKa = 11.84VSRR336 pKa = 11.84LGNRR340 pKa = 11.84GTIRR344 pKa = 11.84TRR346 pKa = 11.84SGTQIGAQVHH356 pKa = 6.09FFQDD360 pKa = 3.77LSSIDD365 pKa = 3.65AEE367 pKa = 4.43EE368 pKa = 4.3GLEE371 pKa = 4.03LALLGQHH378 pKa = 6.43SGDD381 pKa = 3.45ASIVQAQAEE390 pKa = 4.84SVFLGDD396 pKa = 4.51GLHH399 pKa = 7.4PDD401 pKa = 3.42TDD403 pKa = 4.72SNLSLLDD410 pKa = 3.46EE411 pKa = 4.45FAEE414 pKa = 4.71DD415 pKa = 4.6FSHH418 pKa = 6.22SQLIIGEE425 pKa = 4.17RR426 pKa = 11.84KK427 pKa = 9.46ANQVSLPDD435 pKa = 3.84FASPKK440 pKa = 7.8YY441 pKa = 10.58SKK443 pKa = 11.19YY444 pKa = 10.5FLQDD448 pKa = 3.1YY449 pKa = 10.64GNGFIVSHH457 pKa = 6.48PMQTEE462 pKa = 3.92RR463 pKa = 11.84PEE465 pKa = 4.72IIHH468 pKa = 6.95PDD470 pKa = 2.91IDD472 pKa = 3.95AGPTIVIEE480 pKa = 4.58AFDD483 pKa = 3.84SSGGFYY489 pKa = 10.3LHH491 pKa = 7.23PSFVGKK497 pKa = 9.65KK498 pKa = 9.07RR499 pKa = 11.84KK500 pKa = 9.34RR501 pKa = 11.84LYY503 pKa = 10.69FLL505 pKa = 4.65
MM1 pKa = 7.26ARR3 pKa = 11.84SRR5 pKa = 11.84RR6 pKa = 11.84TKK8 pKa = 10.16RR9 pKa = 11.84ASAQDD14 pKa = 3.87LYY16 pKa = 11.2KK17 pKa = 10.69SCLSGGDD24 pKa = 3.86CPPDD28 pKa = 3.34VKK30 pKa = 10.92NKK32 pKa = 10.19YY33 pKa = 7.32EE34 pKa = 4.07QNTTADD40 pKa = 4.17KK41 pKa = 10.18ILKK44 pKa = 9.13YY45 pKa = 10.75GSTAVYY51 pKa = 10.09FGGLGIGTGKK61 pKa = 8.82GTGGVTGYY69 pKa = 8.86TPLGGGEE76 pKa = 4.15GPVRR80 pKa = 11.84VGVPRR85 pKa = 11.84VLRR88 pKa = 11.84PALATDD94 pKa = 4.08LVGPLDD100 pKa = 4.04IAPVDD105 pKa = 3.79AGVGANDD112 pKa = 4.01PSVITLTDD120 pKa = 3.11STLSVDD126 pKa = 3.6VGPGEE131 pKa = 4.26VEE133 pKa = 4.4VVAEE137 pKa = 3.99IHH139 pKa = 6.19PVPDD143 pKa = 3.32TSTTTTGSGQGTSAVLEE160 pKa = 4.22ITPEE164 pKa = 3.79ATPGKK169 pKa = 9.74VRR171 pKa = 11.84VSRR174 pKa = 11.84TQYY177 pKa = 10.34HH178 pKa = 5.77NPAFHH183 pKa = 7.44ILTTSTPIGGEE194 pKa = 3.8TSAVDD199 pKa = 3.42NVFVDD204 pKa = 4.89FGSPSGHH211 pKa = 5.98IVGADD216 pKa = 3.15GVEE219 pKa = 4.34VYY221 pKa = 10.82EE222 pKa = 4.77EE223 pKa = 4.42IPLEE227 pKa = 3.92PLNRR231 pKa = 11.84SEE233 pKa = 5.98FEE235 pKa = 4.32IEE237 pKa = 3.91EE238 pKa = 4.05QTPKK242 pKa = 10.87SSTPKK247 pKa = 10.35ILDD250 pKa = 3.97TIADD254 pKa = 4.16SVRR257 pKa = 11.84KK258 pKa = 9.53FYY260 pKa = 11.07NRR262 pKa = 11.84RR263 pKa = 11.84IQQIKK268 pKa = 7.94TSDD271 pKa = 3.42YY272 pKa = 10.93RR273 pKa = 11.84FLSSPGKK280 pKa = 10.39LVDD283 pKa = 4.01ASFINPAYY291 pKa = 10.59DD292 pKa = 3.68PDD294 pKa = 3.71EE295 pKa = 4.23TLVFDD300 pKa = 3.88TDD302 pKa = 3.43INEE305 pKa = 4.17VQMAPNPDD313 pKa = 3.45FQDD316 pKa = 2.64IRR318 pKa = 11.84VLNRR322 pKa = 11.84PMFSTKK328 pKa = 9.67EE329 pKa = 3.8GYY331 pKa = 9.81IRR333 pKa = 11.84VSRR336 pKa = 11.84LGNRR340 pKa = 11.84GTIRR344 pKa = 11.84TRR346 pKa = 11.84SGTQIGAQVHH356 pKa = 6.09FFQDD360 pKa = 3.77LSSIDD365 pKa = 3.65AEE367 pKa = 4.43EE368 pKa = 4.3GLEE371 pKa = 4.03LALLGQHH378 pKa = 6.43SGDD381 pKa = 3.45ASIVQAQAEE390 pKa = 4.84SVFLGDD396 pKa = 4.51GLHH399 pKa = 7.4PDD401 pKa = 3.42TDD403 pKa = 4.72SNLSLLDD410 pKa = 3.46EE411 pKa = 4.45FAEE414 pKa = 4.71DD415 pKa = 4.6FSHH418 pKa = 6.22SQLIIGEE425 pKa = 4.17RR426 pKa = 11.84KK427 pKa = 9.46ANQVSLPDD435 pKa = 3.84FASPKK440 pKa = 7.8YY441 pKa = 10.58SKK443 pKa = 11.19YY444 pKa = 10.5FLQDD448 pKa = 3.1YY449 pKa = 10.64GNGFIVSHH457 pKa = 6.48PMQTEE462 pKa = 3.92RR463 pKa = 11.84PEE465 pKa = 4.72IIHH468 pKa = 6.95PDD470 pKa = 2.91IDD472 pKa = 3.95AGPTIVIEE480 pKa = 4.58AFDD483 pKa = 3.84SSGGFYY489 pKa = 10.3LHH491 pKa = 7.23PSFVGKK497 pKa = 9.65KK498 pKa = 9.07RR499 pKa = 11.84KK500 pKa = 9.34RR501 pKa = 11.84LYY503 pKa = 10.69FLL505 pKa = 4.65
Molecular weight: 54.47 kDa
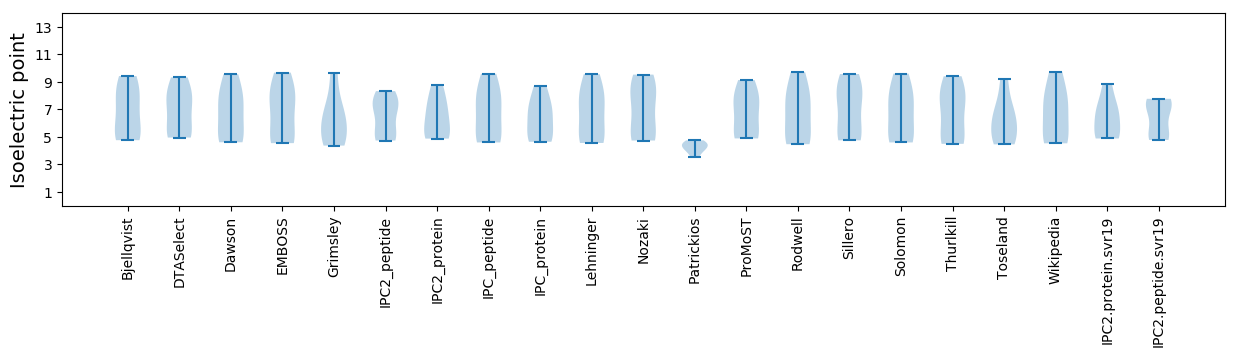
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B7TQP1|B7TQP1_9PAPI E4 (Fragment) OS=Erinaceus europaeus papillomavirus 1 OX=445217 GN=E4 PE=4 SV=1
MM1 pKa = 7.48EE2 pKa = 6.16RR3 pKa = 11.84LTQRR7 pKa = 11.84FDD9 pKa = 3.36ALQEE13 pKa = 3.94KK14 pKa = 10.44LMEE17 pKa = 4.82IYY19 pKa = 10.61EE20 pKa = 4.32KK21 pKa = 10.62DD22 pKa = 3.66SRR24 pKa = 11.84DD25 pKa = 3.6LGVMTSHH32 pKa = 7.53WALQRR37 pKa = 11.84EE38 pKa = 4.6EE39 pKa = 4.09QALLHH44 pKa = 5.55CARR47 pKa = 11.84RR48 pKa = 11.84KK49 pKa = 9.58GVLRR53 pKa = 11.84VGFQPVPPLKK63 pKa = 10.74VSEE66 pKa = 4.44QKK68 pKa = 10.92AKK70 pKa = 10.75AAIEE74 pKa = 3.94MHH76 pKa = 6.32LTLQSLQKK84 pKa = 10.15SAYY87 pKa = 10.21SNEE90 pKa = 3.87DD91 pKa = 2.57WTLSQTSRR99 pKa = 11.84EE100 pKa = 4.19SFMTPPRR107 pKa = 11.84HH108 pKa = 5.78CFKK111 pKa = 10.9KK112 pKa = 9.91QGHH115 pKa = 4.64TVEE118 pKa = 4.25VTFDD122 pKa = 3.74GEE124 pKa = 4.63SANSMLYY131 pKa = 9.2TMWGRR136 pKa = 11.84VYY138 pKa = 10.99YY139 pKa = 10.93QMDD142 pKa = 4.01DD143 pKa = 4.01GSWSVASSGVDD154 pKa = 3.19YY155 pKa = 11.49YY156 pKa = 11.47GIYY159 pKa = 10.85YY160 pKa = 9.91NDD162 pKa = 3.87SEE164 pKa = 4.82GKK166 pKa = 7.3PHH168 pKa = 6.73YY169 pKa = 9.18YY170 pKa = 10.67VKK172 pKa = 10.45FAEE175 pKa = 4.62DD176 pKa = 3.16AQKK179 pKa = 10.97YY180 pKa = 9.54SKK182 pKa = 9.8TGTWFVRR189 pKa = 11.84SNDD192 pKa = 3.47KK193 pKa = 10.4TISAPVTSTCTSPTTRR209 pKa = 11.84DD210 pKa = 3.04RR211 pKa = 11.84SRR213 pKa = 11.84SPRR216 pKa = 11.84RR217 pKa = 11.84HH218 pKa = 5.09TEE220 pKa = 4.02EE221 pKa = 4.12VDD223 pKa = 3.29STTPPGRR230 pKa = 11.84RR231 pKa = 11.84YY232 pKa = 9.67PPSSPPSVSSLRR244 pKa = 11.84LRR246 pKa = 11.84GRR248 pKa = 11.84GGGEE252 pKa = 3.69GEE254 pKa = 4.35YY255 pKa = 10.79YY256 pKa = 10.21PFGRR260 pKa = 11.84SSPSEE265 pKa = 4.08DD266 pKa = 3.06RR267 pKa = 11.84CGSAGSPARR276 pKa = 11.84PVGRR280 pKa = 11.84RR281 pKa = 11.84PPHH284 pKa = 6.45CPQGTPAEE292 pKa = 4.21ASHH295 pKa = 6.54RR296 pKa = 11.84LLWGPRR302 pKa = 11.84DD303 pKa = 3.67PTVLVAKK310 pKa = 10.54GDD312 pKa = 4.14ANTLKK317 pKa = 10.4CWRR320 pKa = 11.84NRR322 pKa = 11.84CRR324 pKa = 11.84KK325 pKa = 6.46THH327 pKa = 6.4AGSFIAFSTTWQWCGDD343 pKa = 3.69GNCRR347 pKa = 11.84QGRR350 pKa = 11.84HH351 pKa = 5.21RR352 pKa = 11.84LQILFSSSQQLDD364 pKa = 3.32QFLGKK369 pKa = 10.54VKK371 pKa = 10.16VPKK374 pKa = 10.09GVEE377 pKa = 3.83VHH379 pKa = 6.96RR380 pKa = 11.84STFDD384 pKa = 3.2GLL386 pKa = 3.71
MM1 pKa = 7.48EE2 pKa = 6.16RR3 pKa = 11.84LTQRR7 pKa = 11.84FDD9 pKa = 3.36ALQEE13 pKa = 3.94KK14 pKa = 10.44LMEE17 pKa = 4.82IYY19 pKa = 10.61EE20 pKa = 4.32KK21 pKa = 10.62DD22 pKa = 3.66SRR24 pKa = 11.84DD25 pKa = 3.6LGVMTSHH32 pKa = 7.53WALQRR37 pKa = 11.84EE38 pKa = 4.6EE39 pKa = 4.09QALLHH44 pKa = 5.55CARR47 pKa = 11.84RR48 pKa = 11.84KK49 pKa = 9.58GVLRR53 pKa = 11.84VGFQPVPPLKK63 pKa = 10.74VSEE66 pKa = 4.44QKK68 pKa = 10.92AKK70 pKa = 10.75AAIEE74 pKa = 3.94MHH76 pKa = 6.32LTLQSLQKK84 pKa = 10.15SAYY87 pKa = 10.21SNEE90 pKa = 3.87DD91 pKa = 2.57WTLSQTSRR99 pKa = 11.84EE100 pKa = 4.19SFMTPPRR107 pKa = 11.84HH108 pKa = 5.78CFKK111 pKa = 10.9KK112 pKa = 9.91QGHH115 pKa = 4.64TVEE118 pKa = 4.25VTFDD122 pKa = 3.74GEE124 pKa = 4.63SANSMLYY131 pKa = 9.2TMWGRR136 pKa = 11.84VYY138 pKa = 10.99YY139 pKa = 10.93QMDD142 pKa = 4.01DD143 pKa = 4.01GSWSVASSGVDD154 pKa = 3.19YY155 pKa = 11.49YY156 pKa = 11.47GIYY159 pKa = 10.85YY160 pKa = 9.91NDD162 pKa = 3.87SEE164 pKa = 4.82GKK166 pKa = 7.3PHH168 pKa = 6.73YY169 pKa = 9.18YY170 pKa = 10.67VKK172 pKa = 10.45FAEE175 pKa = 4.62DD176 pKa = 3.16AQKK179 pKa = 10.97YY180 pKa = 9.54SKK182 pKa = 9.8TGTWFVRR189 pKa = 11.84SNDD192 pKa = 3.47KK193 pKa = 10.4TISAPVTSTCTSPTTRR209 pKa = 11.84DD210 pKa = 3.04RR211 pKa = 11.84SRR213 pKa = 11.84SPRR216 pKa = 11.84RR217 pKa = 11.84HH218 pKa = 5.09TEE220 pKa = 4.02EE221 pKa = 4.12VDD223 pKa = 3.29STTPPGRR230 pKa = 11.84RR231 pKa = 11.84YY232 pKa = 9.67PPSSPPSVSSLRR244 pKa = 11.84LRR246 pKa = 11.84GRR248 pKa = 11.84GGGEE252 pKa = 3.69GEE254 pKa = 4.35YY255 pKa = 10.79YY256 pKa = 10.21PFGRR260 pKa = 11.84SSPSEE265 pKa = 4.08DD266 pKa = 3.06RR267 pKa = 11.84CGSAGSPARR276 pKa = 11.84PVGRR280 pKa = 11.84RR281 pKa = 11.84PPHH284 pKa = 6.45CPQGTPAEE292 pKa = 4.21ASHH295 pKa = 6.54RR296 pKa = 11.84LLWGPRR302 pKa = 11.84DD303 pKa = 3.67PTVLVAKK310 pKa = 10.54GDD312 pKa = 4.14ANTLKK317 pKa = 10.4CWRR320 pKa = 11.84NRR322 pKa = 11.84CRR324 pKa = 11.84KK325 pKa = 6.46THH327 pKa = 6.4AGSFIAFSTTWQWCGDD343 pKa = 3.69GNCRR347 pKa = 11.84QGRR350 pKa = 11.84HH351 pKa = 5.21RR352 pKa = 11.84LQILFSSSQQLDD364 pKa = 3.32QFLGKK369 pKa = 10.54VKK371 pKa = 10.16VPKK374 pKa = 10.09GVEE377 pKa = 3.83VHH379 pKa = 6.96RR380 pKa = 11.84STFDD384 pKa = 3.2GLL386 pKa = 3.71
Molecular weight: 43.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2282 |
54 |
597 |
326.0 |
36.55 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.135 ± 0.148 | 2.323 ± 0.76 |
6.442 ± 0.533 | 5.96 ± 0.422 |
4.426 ± 0.331 | 6.968 ± 0.986 |
1.884 ± 0.306 | 4.426 ± 0.594 |
5.741 ± 0.602 | 8.94 ± 0.996 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.753 ± 0.291 | 3.812 ± 0.931 |
5.434 ± 0.874 | 4.294 ± 0.196 |
6.004 ± 0.786 | 7.581 ± 0.968 |
6.573 ± 0.403 | 6.529 ± 0.382 |
1.446 ± 0.408 | 3.33 ± 0.196 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
