
Ovis aries papillomavirus 3
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Dyokappapapillomavirus; Dyokappapapillomavirus 1
Average proteome isoelectric point is 5.69
Get precalculated fractions of proteins
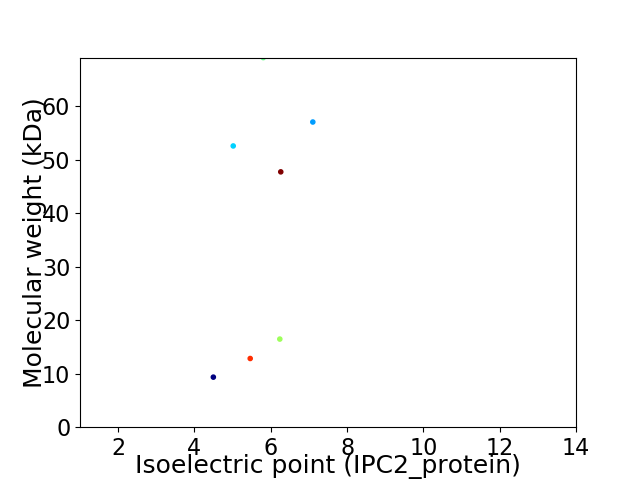
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
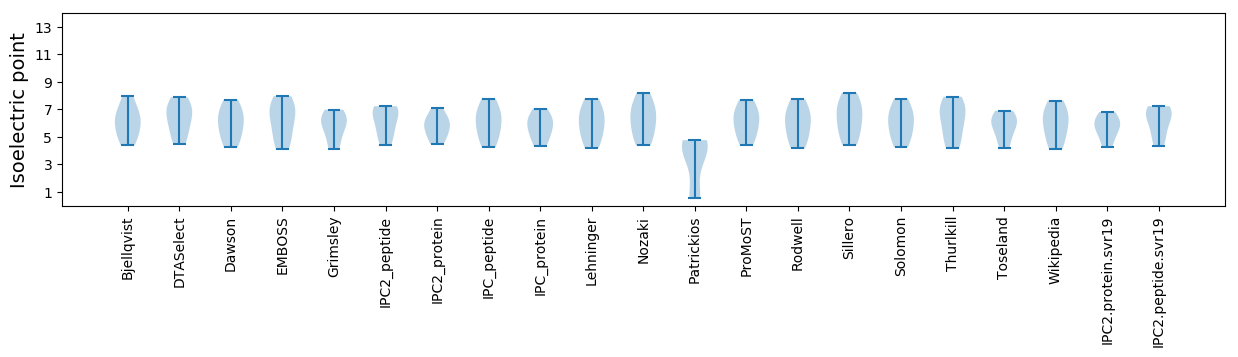
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D5FL27|D5FL27_9PAPI Replication protein E1 OS=Ovis aries papillomavirus 3 OX=634772 GN=E1 PE=3 SV=1
MM1 pKa = 8.07RR2 pKa = 11.84GQQPTLPDD10 pKa = 3.28IVLHH14 pKa = 5.93EE15 pKa = 4.31VPEE18 pKa = 4.3PVSLYY23 pKa = 10.5CDD25 pKa = 3.35EE26 pKa = 4.14QLEE29 pKa = 4.4EE30 pKa = 4.63EE31 pKa = 4.56EE32 pKa = 4.58EE33 pKa = 4.24QPALYY38 pKa = 9.49TLYY41 pKa = 9.52TLCGRR46 pKa = 11.84CCQRR50 pKa = 11.84LKK52 pKa = 10.18LTFRR56 pKa = 11.84AFTQTKK62 pKa = 8.05ITLEE66 pKa = 4.0TLLLGDD72 pKa = 5.39LDD74 pKa = 5.14LFCPPCAAA82 pKa = 4.79
MM1 pKa = 8.07RR2 pKa = 11.84GQQPTLPDD10 pKa = 3.28IVLHH14 pKa = 5.93EE15 pKa = 4.31VPEE18 pKa = 4.3PVSLYY23 pKa = 10.5CDD25 pKa = 3.35EE26 pKa = 4.14QLEE29 pKa = 4.4EE30 pKa = 4.63EE31 pKa = 4.56EE32 pKa = 4.58EE33 pKa = 4.24QPALYY38 pKa = 9.49TLYY41 pKa = 9.52TLCGRR46 pKa = 11.84CCQRR50 pKa = 11.84LKK52 pKa = 10.18LTFRR56 pKa = 11.84AFTQTKK62 pKa = 8.05ITLEE66 pKa = 4.0TLLLGDD72 pKa = 5.39LDD74 pKa = 5.14LFCPPCAAA82 pKa = 4.79
Molecular weight: 9.36 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D8L564|D8L564_9PAPI E4 OS=Ovis aries papillomavirus 3 OX=634772 PE=4 SV=1
MM1 pKa = 7.28AVWVPNGKK9 pKa = 9.27SFYY12 pKa = 10.04LPQSAVTRR20 pKa = 11.84ILSTNEE26 pKa = 3.52YY27 pKa = 9.16VQGTGLVFHH36 pKa = 6.96GSSNRR41 pKa = 11.84LLCVGHH47 pKa = 6.9PFYY50 pKa = 10.17EE51 pKa = 4.37TQRR54 pKa = 11.84PDD56 pKa = 3.18GSVKK60 pKa = 9.91VPKK63 pKa = 10.3VSSSQYY69 pKa = 10.37RR70 pKa = 11.84VFKK73 pKa = 11.09VLLPDD78 pKa = 3.88PNKK81 pKa = 10.18FVFSEE86 pKa = 4.19PNLYY90 pKa = 10.63DD91 pKa = 3.87PEE93 pKa = 4.28SQRR96 pKa = 11.84LVWKK100 pKa = 10.51LRR102 pKa = 11.84GLQVDD107 pKa = 3.96RR108 pKa = 11.84GQPHH112 pKa = 5.92GVWVTGHH119 pKa = 7.18LLMNKK124 pKa = 9.87LDD126 pKa = 3.72DD127 pKa = 3.97TEE129 pKa = 4.15NLGRR133 pKa = 11.84QGSDD137 pKa = 2.75PAGRR141 pKa = 11.84DD142 pKa = 2.73KK143 pKa = 11.18DD144 pKa = 3.74SRR146 pKa = 11.84VNMGLEE152 pKa = 4.18PKK154 pKa = 10.2QMQVLIVGCRR164 pKa = 11.84PPWGEE169 pKa = 3.46HH170 pKa = 3.89WGVAKK175 pKa = 10.5KK176 pKa = 10.25CASDD180 pKa = 3.51NADD183 pKa = 3.67PDD185 pKa = 3.52KK186 pKa = 11.44CPAIEE191 pKa = 4.26LKK193 pKa = 9.63STIIEE198 pKa = 4.84DD199 pKa = 3.88GNMMDD204 pKa = 3.83TGFGNLDD211 pKa = 3.34FRR213 pKa = 11.84SLQEE217 pKa = 4.11NKK219 pKa = 10.11ADD221 pKa = 3.87APIDD225 pKa = 3.45ICQSICKK232 pKa = 9.64YY233 pKa = 10.18PDD235 pKa = 4.81FIRR238 pKa = 11.84MSQEE242 pKa = 3.74TYY244 pKa = 9.06GDD246 pKa = 3.5HH247 pKa = 6.82MFFCAKK253 pKa = 10.17HH254 pKa = 4.23EE255 pKa = 4.23QIYY258 pKa = 9.73LRR260 pKa = 11.84HH261 pKa = 5.69YY262 pKa = 9.56FSKK265 pKa = 10.5AGKK268 pKa = 9.71IGEE271 pKa = 4.39EE272 pKa = 4.35VPKK275 pKa = 9.84TLYY278 pKa = 10.62VPPQPDD284 pKa = 3.68TVNGTVNFWGSPSGSMVSSNNQLFNKK310 pKa = 9.08PYY312 pKa = 9.77WVRR315 pKa = 11.84QAQGHH320 pKa = 5.67NNGVLWNNLAFITVGDD336 pKa = 3.98TTRR339 pKa = 11.84GTNFNISVLDD349 pKa = 4.12NGAQPYY355 pKa = 9.88KK356 pKa = 10.24DD357 pKa = 3.14SSYY360 pKa = 11.78AEE362 pKa = 4.21FLRR365 pKa = 11.84HH366 pKa = 5.81VEE368 pKa = 4.1EE369 pKa = 4.82FDD371 pKa = 3.12IQIIVEE377 pKa = 4.1ACIVDD382 pKa = 4.23LTPEE386 pKa = 3.88IVSFIHH392 pKa = 6.7QMDD395 pKa = 3.82PTILDD400 pKa = 3.32NWNLGIQAAPDD411 pKa = 3.31SSLWEE416 pKa = 4.06TYY418 pKa = 10.12RR419 pKa = 11.84YY420 pKa = 9.62ISSFATKK427 pKa = 10.52CPDD430 pKa = 3.29QVPKK434 pKa = 10.78PEE436 pKa = 4.33APKK439 pKa = 10.74DD440 pKa = 3.7PYY442 pKa = 10.98EE443 pKa = 4.64KK444 pKa = 10.74LSFWTVDD451 pKa = 3.58LNEE454 pKa = 5.17KK455 pKa = 10.28LSQDD459 pKa = 3.43LTHH462 pKa = 6.81FPLGRR467 pKa = 11.84RR468 pKa = 11.84YY469 pKa = 10.06LFQYY473 pKa = 7.15TVRR476 pKa = 11.84PPKK479 pKa = 10.48SAVKK483 pKa = 10.32RR484 pKa = 11.84KK485 pKa = 9.1AANNSGLNSSKK496 pKa = 10.35RR497 pKa = 11.84RR498 pKa = 11.84RR499 pKa = 11.84KK500 pKa = 10.11LNNKK504 pKa = 8.86
MM1 pKa = 7.28AVWVPNGKK9 pKa = 9.27SFYY12 pKa = 10.04LPQSAVTRR20 pKa = 11.84ILSTNEE26 pKa = 3.52YY27 pKa = 9.16VQGTGLVFHH36 pKa = 6.96GSSNRR41 pKa = 11.84LLCVGHH47 pKa = 6.9PFYY50 pKa = 10.17EE51 pKa = 4.37TQRR54 pKa = 11.84PDD56 pKa = 3.18GSVKK60 pKa = 9.91VPKK63 pKa = 10.3VSSSQYY69 pKa = 10.37RR70 pKa = 11.84VFKK73 pKa = 11.09VLLPDD78 pKa = 3.88PNKK81 pKa = 10.18FVFSEE86 pKa = 4.19PNLYY90 pKa = 10.63DD91 pKa = 3.87PEE93 pKa = 4.28SQRR96 pKa = 11.84LVWKK100 pKa = 10.51LRR102 pKa = 11.84GLQVDD107 pKa = 3.96RR108 pKa = 11.84GQPHH112 pKa = 5.92GVWVTGHH119 pKa = 7.18LLMNKK124 pKa = 9.87LDD126 pKa = 3.72DD127 pKa = 3.97TEE129 pKa = 4.15NLGRR133 pKa = 11.84QGSDD137 pKa = 2.75PAGRR141 pKa = 11.84DD142 pKa = 2.73KK143 pKa = 11.18DD144 pKa = 3.74SRR146 pKa = 11.84VNMGLEE152 pKa = 4.18PKK154 pKa = 10.2QMQVLIVGCRR164 pKa = 11.84PPWGEE169 pKa = 3.46HH170 pKa = 3.89WGVAKK175 pKa = 10.5KK176 pKa = 10.25CASDD180 pKa = 3.51NADD183 pKa = 3.67PDD185 pKa = 3.52KK186 pKa = 11.44CPAIEE191 pKa = 4.26LKK193 pKa = 9.63STIIEE198 pKa = 4.84DD199 pKa = 3.88GNMMDD204 pKa = 3.83TGFGNLDD211 pKa = 3.34FRR213 pKa = 11.84SLQEE217 pKa = 4.11NKK219 pKa = 10.11ADD221 pKa = 3.87APIDD225 pKa = 3.45ICQSICKK232 pKa = 9.64YY233 pKa = 10.18PDD235 pKa = 4.81FIRR238 pKa = 11.84MSQEE242 pKa = 3.74TYY244 pKa = 9.06GDD246 pKa = 3.5HH247 pKa = 6.82MFFCAKK253 pKa = 10.17HH254 pKa = 4.23EE255 pKa = 4.23QIYY258 pKa = 9.73LRR260 pKa = 11.84HH261 pKa = 5.69YY262 pKa = 9.56FSKK265 pKa = 10.5AGKK268 pKa = 9.71IGEE271 pKa = 4.39EE272 pKa = 4.35VPKK275 pKa = 9.84TLYY278 pKa = 10.62VPPQPDD284 pKa = 3.68TVNGTVNFWGSPSGSMVSSNNQLFNKK310 pKa = 9.08PYY312 pKa = 9.77WVRR315 pKa = 11.84QAQGHH320 pKa = 5.67NNGVLWNNLAFITVGDD336 pKa = 3.98TTRR339 pKa = 11.84GTNFNISVLDD349 pKa = 4.12NGAQPYY355 pKa = 9.88KK356 pKa = 10.24DD357 pKa = 3.14SSYY360 pKa = 11.78AEE362 pKa = 4.21FLRR365 pKa = 11.84HH366 pKa = 5.81VEE368 pKa = 4.1EE369 pKa = 4.82FDD371 pKa = 3.12IQIIVEE377 pKa = 4.1ACIVDD382 pKa = 4.23LTPEE386 pKa = 3.88IVSFIHH392 pKa = 6.7QMDD395 pKa = 3.82PTILDD400 pKa = 3.32NWNLGIQAAPDD411 pKa = 3.31SSLWEE416 pKa = 4.06TYY418 pKa = 10.12RR419 pKa = 11.84YY420 pKa = 9.62ISSFATKK427 pKa = 10.52CPDD430 pKa = 3.29QVPKK434 pKa = 10.78PEE436 pKa = 4.33APKK439 pKa = 10.74DD440 pKa = 3.7PYY442 pKa = 10.98EE443 pKa = 4.64KK444 pKa = 10.74LSFWTVDD451 pKa = 3.58LNEE454 pKa = 5.17KK455 pKa = 10.28LSQDD459 pKa = 3.43LTHH462 pKa = 6.81FPLGRR467 pKa = 11.84RR468 pKa = 11.84YY469 pKa = 10.06LFQYY473 pKa = 7.15TVRR476 pKa = 11.84PPKK479 pKa = 10.48SAVKK483 pKa = 10.32RR484 pKa = 11.84KK485 pKa = 9.1AANNSGLNSSKK496 pKa = 10.35RR497 pKa = 11.84RR498 pKa = 11.84RR499 pKa = 11.84KK500 pKa = 10.11LNNKK504 pKa = 8.86
Molecular weight: 57.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2362 |
82 |
607 |
337.4 |
37.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.588 ± 0.507 | 2.456 ± 0.618 |
5.8 ± 0.567 | 6.732 ± 0.84 |
3.683 ± 0.508 | 6.647 ± 0.938 |
2.244 ± 0.184 | 3.895 ± 0.361 |
5.207 ± 0.745 | 8.552 ± 0.591 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.651 ± 0.269 | 4.149 ± 0.617 |
6.774 ± 0.829 | 4.742 ± 0.552 |
5.97 ± 0.59 | 7.917 ± 0.763 |
6.181 ± 0.742 | 6.901 ± 0.664 |
1.82 ± 0.345 | 3.091 ± 0.285 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
